8OG8
 
 | | Crystal structure of human DCAF1 WD40 repeats (Q1250L) in complex with compound 3 | | Descriptor: | 1,2-ETHANEDIOL, 5-(2-methyl-1-phenyl-propan-2-yl)imidazo[2,1-a]isoquinoline, ACETATE ION, ... | | Authors: | Schroeder, M, Vulpetti, A, Renatus, M. | | Deposit date: | 2023-03-19 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Discovery of New Binders for DCAF1, an Emerging Ligase Target in the Targeted Protein Degradation Field.
Acs Med.Chem.Lett., 14, 2023
|
|
8OG6
 
 | | Crystal structure of human DCAF1 WD40 repeats (Q1250L) in complex with compound 1 | | Descriptor: | 5-(2-fluorophenyl)-2,3-dihydroimidazo[2,1-a]isoquinoline, ACETATE ION, DDB1- and CUL4-associated factor 1 | | Authors: | Schroeder, M, Vulpetti, A, Renatus, M. | | Deposit date: | 2023-03-19 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.245 Å) | | Cite: | Discovery of New Binders for DCAF1, an Emerging Ligase Target in the Targeted Protein Degradation Field.
Acs Med.Chem.Lett., 14, 2023
|
|
8OG9
 
 | | Crystal structure of human DCAF1 WD40 repeats (Q1250L) in complex with compound 4 | | Descriptor: | 5-[1-(4-chlorophenyl)cyclopropyl]imidazo[2,1-a]isoquinoline, DDB1- and CUL4-associated factor 1 | | Authors: | Schroeder, M, Vulpetti, A, Renatus, M. | | Deposit date: | 2023-03-19 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.945 Å) | | Cite: | Discovery of New Binders for DCAF1, an Emerging Ligase Target in the Targeted Protein Degradation Field.
Acs Med.Chem.Lett., 14, 2023
|
|
8OGB
 
 | | Crystal structure of human DCAF1 WD40 repeats (Q1250L) in complex with compound 8 | | Descriptor: | 1,2-ETHANEDIOL, DDB1- and CUL4-associated factor 1, DIMETHYL SULFOXIDE, ... | | Authors: | Schroeder, M, Vulpetti, A, Renatus, M. | | Deposit date: | 2023-03-19 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Discovery of New Binders for DCAF1, an Emerging Ligase Target in the Targeted Protein Degradation Field.
Acs Med.Chem.Lett., 14, 2023
|
|
8OGC
 
 | | Crystal structure of human DCAF1 WD40 repeats (Q1250L) in complex with compound 11 | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-[4-(2-azanylethylamino)-2-[1-(4-chlorophenyl)cyclopentyl]quinazolin-7-yl]piperazin-1-yl]ethanone, ACETATE ION, ... | | Authors: | Schroeder, M, Vulpetti, A, Renatus, M. | | Deposit date: | 2023-03-19 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Discovery of New Binders for DCAF1, an Emerging Ligase Target in the Targeted Protein Degradation Field.
Acs Med.Chem.Lett., 14, 2023
|
|
8OG7
 
 | | Crystal structure of human DCAF1 WD40 repeats (Q1250L) in complex with compound 2 | | Descriptor: | (4~{R})-4-[3-(4-chloranylphenoxy)phenyl]pyrrolidin-2-imine, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Schroeder, M, Vulpetti, A, Renatus, M. | | Deposit date: | 2023-03-19 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Discovery of New Binders for DCAF1, an Emerging Ligase Target in the Targeted Protein Degradation Field.
Acs Med.Chem.Lett., 14, 2023
|
|
8OGA
 
 | | Crystal structure of human DCAF1 WD40 repeats (Q1250L) in complex with compound 6 | | Descriptor: | 1,2-ETHANEDIOL, 5-[1-(4-methoxyphenyl)cyclopropyl]-8-(4-methylpiperazin-1-yl)-2,3-dihydroimidazo[2,1-a]isoquinoline, DDB1- and CUL4-associated factor 1, ... | | Authors: | Schroeder, M, Vulpetti, A, Renatus, M. | | Deposit date: | 2023-03-19 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of New Binders for DCAF1, an Emerging Ligase Target in the Targeted Protein Degradation Field.
Acs Med.Chem.Lett., 14, 2023
|
|
6YWL
 
 | | Crystal structure of SARS-CoV-2 (Covid-19) NSP3 macrodomain in complex with ADP-ribose | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5-DIPHOSPHORIBOSE, MAGNESIUM ION, ... | | Authors: | Schroeder, M, Ni, X, Olieric, V, Sharpe, E.M, Wojdyla, J.A, Wang, M, Knapp, S, Chaikuad, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-29 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insights into Plasticity and Discovery of Remdesivir Metabolite GS-441524 Binding in SARS-CoV-2 Macrodomain.
Acs Med.Chem.Lett., 12, 2021
|
|
8OOD
 
 | |
8OO5
 
 | |
6YTD
 
 | | CLK1 V324A mutant bound with benzothiazole Tg003 (Cpd 2) | | Descriptor: | (1~{Z})-1-(3-ethyl-5-methoxy-1,3-benzothiazol-2-ylidene)propan-2-one, Dual specificity protein kinase CLK1 | | Authors: | Schroeder, M, Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-24 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | DFG-1 Residue Controls Inhibitor Binding Mode and Affinity, Providing a Basis for Rational Design of Kinase Inhibitor Selectivity.
J.Med.Chem., 63, 2020
|
|
6YTI
 
 | | CLK1 bound with ETH1610 (Cpd 17) | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity protein kinase CLK1, methyl 9-[(2-fluoranyl-4-methoxy-phenyl)amino]-[1,3]thiazolo[5,4-f]quinazoline-2-carboximidate | | Authors: | Schroeder, M, Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-24 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | DFG-1 Residue Controls Inhibitor Binding Mode and Affinity, Providing a Basis for Rational Design of Kinase Inhibitor Selectivity.
J.Med.Chem., 63, 2020
|
|
6YU1
 
 | | CLK3 bound with beta-carboline KH-CARB13 (Cpd 3) | | Descriptor: | (4~{S})-7,8-bis(chloranyl)-9-methyl-1-oxidanylidene-spiro[2,4-dihydropyrido[3,4-b]indole-3,4'-piperidine]-4-carbonitrile, 1,2-ETHANEDIOL, Dual specificity protein kinase CLK3, ... | | Authors: | Schroeder, M, Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-25 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | DFG-1 Residue Controls Inhibitor Binding Mode and Affinity, Providing a Basis for Rational Design of Kinase Inhibitor Selectivity.
J.Med.Chem., 63, 2020
|
|
6YTA
 
 | | CLK1 bound with imidazopyridazine (Cpd 1) | | Descriptor: | 1,2-ETHANEDIOL, 1-(3-{6-[(CYCLOPROPYLMETHYL)AMINO]IMIDAZO[1,2-B]PYRIDAZIN-3-YL}PHENYL)ETHANONE, Dual specificity protein kinase CLK1 | | Authors: | Schroeder, M, Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-24 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DFG-1 Residue Controls Inhibitor Binding Mode and Affinity, Providing a Basis for Rational Design of Kinase Inhibitor Selectivity.
J.Med.Chem., 63, 2020
|
|
6YTG
 
 | | CLK1 bound with beta-carboline KH-CARB13 (Cpd 3) | | Descriptor: | (4~{S})-7,8-bis(chloranyl)-9-methyl-1-oxidanylidene-spiro[2,4-dihydropyrido[3,4-b]indole-3,4'-piperidine]-4-carbonitrile, Dual specificity protein kinase CLK1 | | Authors: | Schroeder, M, Chaikuad, A, Huber, K, Bracher, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-24 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | DFG-1 Residue Controls Inhibitor Binding Mode and Affinity, Providing a Basis for Rational Design of Kinase Inhibitor Selectivity.
J.Med.Chem., 63, 2020
|
|
6YTW
 
 | | CLK3 bound with benzothiazole Tg003 (Cpd 2) | | Descriptor: | (1~{Z})-1-(3-ethyl-5-methoxy-1,3-benzothiazol-2-ylidene)propan-2-one, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Schroeder, M, Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-24 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | DFG-1 Residue Controls Inhibitor Binding Mode and Affinity, Providing a Basis for Rational Design of Kinase Inhibitor Selectivity.
J.Med.Chem., 63, 2020
|
|
6YTY
 
 | | CLK3 A319V mutant bound with benzothiazole Tg003 (Cpd 2) | | Descriptor: | (1~{Z})-1-(3-ethyl-5-methoxy-1,3-benzothiazol-2-ylidene)propan-2-one, 1,2-ETHANEDIOL, Dual specificity protein kinase CLK3 | | Authors: | Schroeder, M, Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-24 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | DFG-1 Residue Controls Inhibitor Binding Mode and Affinity, Providing a Basis for Rational Design of Kinase Inhibitor Selectivity.
J.Med.Chem., 63, 2020
|
|
6YTE
 
 | | CLK1 bound with benzothiazole Tg003 (Cpd 2) | | Descriptor: | (1~{Z})-1-(3-ethyl-5-methoxy-1,3-benzothiazol-2-ylidene)propan-2-one, Dual specificity protein kinase CLK1 | | Authors: | Schroeder, M, Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-24 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DFG-1 Residue Controls Inhibitor Binding Mode and Affinity, Providing a Basis for Rational Design of Kinase Inhibitor Selectivity.
J.Med.Chem., 63, 2020
|
|
6ZLN
 
 | | CLK1 bound with GW807982X (Cpd 8) | | Descriptor: | 1,2-ETHANEDIOL, 4-(6-ethoxypyrazolo[1,5-b]pyridazin-3-yl)-~{N}-[3-methoxy-5-(trifluoromethyl)phenyl]pyrimidin-2-amine, Dual specificity protein kinase CLK1 | | Authors: | Schroeder, M, Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-06-30 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | DFG-1 Residue Controls Inhibitor Binding Mode and Affinity, Providing a Basis for Rational Design of Kinase Inhibitor Selectivity.
J.Med.Chem., 63, 2020
|
|
6Z2V
 
 | | CLK3 A319V mutant bound with beta-carboline KH-CARB13 (Cpd 3) | | Descriptor: | (4~{S})-7,8-bis(chloranyl)-9-methyl-1-oxidanylidene-spiro[2,4-dihydropyrido[3,4-b]indole-3,4'-piperidine]-4-carbonitrile, 1,2-ETHANEDIOL, Dual specificity protein kinase CLK3, ... | | Authors: | Schroeder, M, Chaikuad, A, Bracher, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-05-18 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | DFG-1 Residue Controls Inhibitor Binding Mode and Affinity, Providing a Basis for Rational Design of Kinase Inhibitor Selectivity.
J.Med.Chem., 63, 2020
|
|
7AK3
 
 | | CLK1 bound with CAF052 | | Descriptor: | Dual specificity protein kinase CLK1, ~{N}-[3-fluoranyl-4-(4-methylpiperazin-1-yl)phenyl]-4-pyrazolo[1,5-b]pyridazin-3-yl-pyrimidin-2-amine | | Authors: | Schroeder, M, Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-09-29 | | Release date: | 2020-11-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure and Inhibitor Identifications Reveal Targeting Opportunity for the Atypical MAPK Kinase ERK3.
Int J Mol Sci, 21, 2020
|
|
1QAD
 
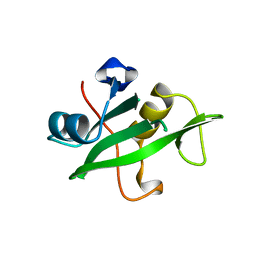 | | Crystal Structure of the C-Terminal SH2 Domain of the P85 alpha Regulatory Subunit of Phosphoinositide 3-Kinase: An SH2 domain mimicking its own substrate | | Descriptor: | PI3-KINASE P85 ALPHA SUBUNIT | | Authors: | Hoedemaeker, P.J, Siegal, G, Roe, M, Driscoll, P.C, Abrahams, J.P.A. | | Deposit date: | 1999-02-26 | | Release date: | 1999-10-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the C-terminal SH2 domain of the p85alpha regulatory subunit of phosphoinositide 3-kinase: an SH2 domain mimicking its own substrate.
J.Mol.Biol., 292, 1999
|
|
2PRN
 
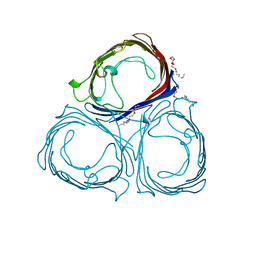 | | RHODOPSEUDOMONAS BLASTICA PORIN, TRIPLE MUTANT E1M, E99W, A116W | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, MAGNESIUM ION, PORIN | | Authors: | Maveyraud, L, Schmid, B, Schulz, G.E. | | Deposit date: | 1998-06-12 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Porin mutants with new channel properties.
Protein Sci., 7, 1998
|
|
8DUA
 
 | | SIV E660.CR54 SOS-2P Env Trimer with ITS92.02 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, Envelope glycoprotein gp41, ... | | Authors: | Gorman, J, Kwong, P.D. | | Deposit date: | 2022-07-27 | | Release date: | 2022-10-26 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.32 Å) | | Cite: | Cryo-EM structures of prefusion SIV envelope trimer.
Nat.Struct.Mol.Biol., 29, 2022
|
|
8DVD
 
 | |
