8THQ
 
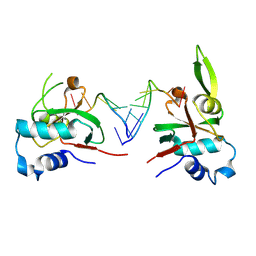 | | Nonamer RNA bound to hAgo2-PAZ | | Descriptor: | Protein argonaute-2, RNA (5'-R(*CP*GP*UP*GP*AP*CP*UP*CP*U)-3') | | Authors: | Pallan, P.S, Harp, J.M, Egli, M. | | Deposit date: | 2023-07-17 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Single-Stranded Hairpin Loop RNAs (loopmeRNAs) Potently Induce Gene Silencing through the RNA Interference Pathway.
J.Am.Chem.Soc., 2024
|
|
8EQQ
 
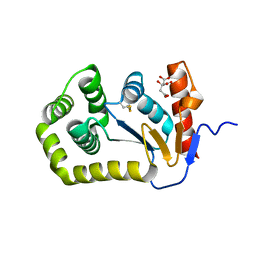 | | Crystal structure of E.coli DsbA mutant E37A | | Descriptor: | CITRATE ANION, Thiol:disulfide interchange protein DsbA | | Authors: | Wang, G, Heras, B. | | Deposit date: | 2022-10-09 | | Release date: | 2023-02-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | A Buried Water Network Modulates the Activity of the Escherichia coli Disulphide Catalyst DsbA.
Antioxidants, 12, 2023
|
|
8EOC
 
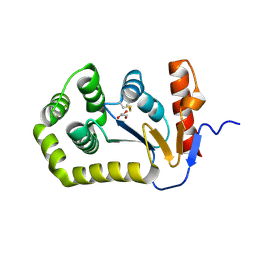 | | Crystal structure of E.coli DsbA mutant E24A/K58A | | Descriptor: | COPPER (II) ION, GLYCEROL, Thiol:disulfide interchange protein DsbA | | Authors: | Wang, G, Heras, B. | | Deposit date: | 2022-10-03 | | Release date: | 2023-02-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | A Buried Water Network Modulates the Activity of the Escherichia coli Disulphide Catalyst DsbA.
Antioxidants, 12, 2023
|
|
8EQO
 
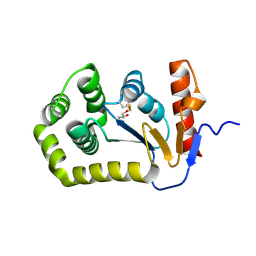 | | Crystal structure of E.coli DsbA mutant K58A | | Descriptor: | COPPER (II) ION, GLYCEROL, Thiol:disulfide interchange protein DsbA | | Authors: | Wang, G, Heras, B. | | Deposit date: | 2022-10-08 | | Release date: | 2023-02-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | A Buried Water Network Modulates the Activity of the Escherichia coli Disulphide Catalyst DsbA.
Antioxidants, 12, 2023
|
|
8EQP
 
 | | Crystal structure of E.coli DsbA mutant E24A/E37A/K58A | | Descriptor: | CITRATE ANION, GLYCEROL, Thiol:disulfide interchange protein DsbA | | Authors: | Wang, G, Heras, B. | | Deposit date: | 2022-10-09 | | Release date: | 2023-02-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Buried Water Network Modulates the Activity of the Escherichia coli Disulphide Catalyst DsbA.
Antioxidants, 12, 2023
|
|
8EQR
 
 | | Crystal structure of E.coli DsbA mutant E24A | | Descriptor: | DI(HYDROXYETHYL)ETHER, Thiol:disulfide interchange protein DsbA | | Authors: | Wang, G, Heras, B. | | Deposit date: | 2022-10-09 | | Release date: | 2023-02-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | A Buried Water Network Modulates the Activity of the Escherichia coli Disulphide Catalyst DsbA.
Antioxidants, 12, 2023
|
|
2KLK
 
 | | Solution structure of GB1 A34F mutant with RDC and SAXS | | Descriptor: | IMMUNOGLOBULIN G-BINDING PROTEIN G | | Authors: | Wang, J, Zuo, X, Yu, P, Byeon, I.L, Jung, J, Schwieters, C.D, Gronenborn, A.M, Wang, Y. | | Deposit date: | 2009-07-06 | | Release date: | 2009-10-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Determination of multicomponent protein structures in solution using global orientation and shape restraints.
J.Am.Chem.Soc., 131, 2009
|
|
2LKO
 
 | | Structural Basis of Phosphoinositide Binding to Kindlin-2 Pleckstrin Homology Domain in Regulating Integrin Activation | | Descriptor: | Fermitin family homolog 2, INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE | | Authors: | Liu, J, Fukuda, K, Xu, Z. | | Deposit date: | 2011-10-17 | | Release date: | 2011-10-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of phosphoinositide binding to kindlin-2 protein pleckstrin homology domain in regulating integrin activation.
J.Biol.Chem., 286, 2011
|
|
2F42
 
 | |
2MSU
 
 | |
5Y9V
 
 | | Crystal structure of diamondback moth ryanodine receptor N-terminal domain | | Descriptor: | CHLORIDE ION, GLYCEROL, Ryanodine receptor 1 | | Authors: | Lin, L, Yuchi, Z. | | Deposit date: | 2017-08-28 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.841 Å) | | Cite: | Crystal structure of ryanodine receptor N-terminal domain from Plutella xylostella reveals two potential species-specific insecticide-targeting sites.
Insect Biochem. Mol. Biol., 92, 2017
|
|
4FS8
 
 | | The structure of an As(III) S-adenosylmethionine methyltransferase: insights into the mechanism of arsenic biotransformation | | Descriptor: | Arsenic methyltransferase, CALCIUM ION | | Authors: | Ajees, A.A, Marapakala, K, Packianathan, C, Sankaran, B, Rosen, B.P. | | Deposit date: | 2012-06-27 | | Release date: | 2012-07-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure of an As(III) S-Adenosylmethionine Methyltransferase: Insights into the Mechanism of Arsenic Biotransformation.
Biochemistry, 51, 2012
|
|
4F7H
 
 | | The crystal structure of kindlin-2 pleckstrin homology domain in free form | | Descriptor: | Fermitin family homolog 2, S,R MESO-TARTARIC ACID | | Authors: | Liu, Y, Zhu, Y, Qin, J, Ye, S, Zhang, R. | | Deposit date: | 2012-05-16 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of kindlin-2 PH domain reveals a conformational transition for its membrane anchoring and regulation of integrin activation.
Protein Cell, 3, 2012
|
|
8G6F
 
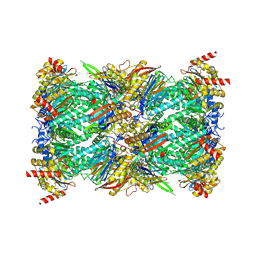 | | Structure of the Plasmodium falciparum 20S proteasome beta-6 A117D mutant complexed with inhibitor WLW-vs | | Descriptor: | (2S)-N-[(E,2S)-1-(1H-indol-3-yl)-4-methylsulfonyl-but-3-en-2-yl]-2-[[(2S)-3-(1H-indol-3-yl)-2-(2-morpholin-4-ylethanoylamino)propanoyl]amino]-4-methyl-pentanamide, Proteasome endopeptidase complex, Proteasome subunit alpha type-1, ... | | Authors: | Hsu, H.-C, Li, H. | | Deposit date: | 2023-02-15 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Structures revealing mechanisms of resistance and collateral sensitivity of Plasmodium falciparum to proteasome inhibitors.
Nat Commun, 14, 2023
|
|
8G6E
 
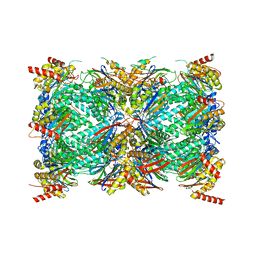 | | Structure of the Plasmodium falciparum 20S proteasome complexed with inhibitor TDI-8304 | | Descriptor: | (7S,10S,13S)-N-cyclopentyl-10-[2-(morpholin-4-yl)ethyl]-9,12-dioxo-13-(2-oxopyrrolidin-1-yl)-2-oxa-8,11-diazabicyclo[13.3.1]nonadeca-1(19),15,17-triene-7-carboxamide, Proteasome subunit alpha type, Proteasome subunit alpha type-1, ... | | Authors: | Hsu, H.-C, Li, H. | | Deposit date: | 2023-02-15 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (2.18 Å) | | Cite: | Structures revealing mechanisms of resistance and collateral sensitivity of Plasmodium falciparum to proteasome inhibitors.
Nat Commun, 14, 2023
|
|
8IKL
 
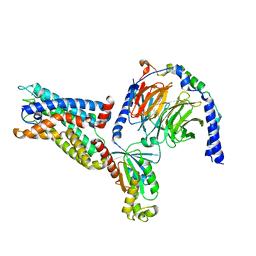 | | Cryo-EM structure of the CD97-G13 complex | | Descriptor: | Adhesion G protein-coupled receptor E5, Guanine nucleotide-binding protein G(13) subunit alpha isoforms short, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Mao, C, Zhao, R, Dong, Y, Gao, M, Chen, L, Zhang, C, Xiao, P. | | Deposit date: | 2023-02-28 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Conformational transitions and activation of the adhesion receptor CD97.
Mol.Cell, 84, 2024
|
|
3P4A
 
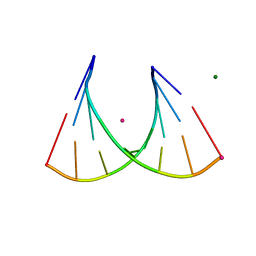 | | 2'Fluoro modified RNA octamer fA2U2 | | Descriptor: | 2'Fluoro modified RNA 8-MER, MAGNESIUM ION, STRONTIUM ION | | Authors: | Manoharan, M, Akinc, A, Pandey, R.K, Qin, J, Hadwiger, P, John, M, Mills, K, Charisse, K, Maier, M.A, Nechev, L, Greene, E.M, Pallan, P.S, Rozners, E, Rajeev, K.G, Egli, M. | | Deposit date: | 2010-10-06 | | Release date: | 2011-01-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Unexpected origins of the enhanced pairing affinity of 2'-fluoro-modified RNA.
Nucleic Acids Res., 39, 2011
|
|
8IA7
 
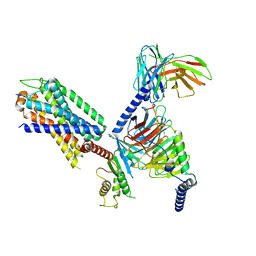 | | Structural insights into human brain gut peptide cholecystokinin receptors | | Descriptor: | CCK-8, Gastrin/cholecystokinin type B receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Ding, Y, Zhang, H, Liao, Y, Chen, L, Ji, S. | | Deposit date: | 2023-02-08 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into human brain-gut peptide cholecystokinin receptors.
Cell Discov, 8, 2022
|
|
6URC
 
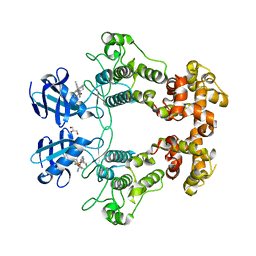 | | Crystal structure of IRE1a in complex with compound 18 | | Descriptor: | 2-chloro-N-(6-methyl-5-{[3-(2-{[(3S)-piperidin-3-yl]amino}pyrimidin-4-yl)pyridin-2-yl]oxy}naphthalen-1-yl)benzene-1-sulfonamide, GLYCEROL, Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Wallweber, H.H, Wang, W. | | Deposit date: | 2019-10-23 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Disruption of IRE1 alpha through its kinase domain attenuates multiple myeloma.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
8GA8
 
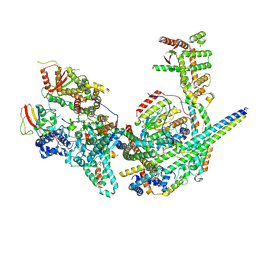 | | Structure of the yeast (HDAC) Rpd3L complex | | Descriptor: | Histone deacetylase RPD3, Transcriptional regulatory protein DEP1, Transcriptional regulatory protein PHO23, ... | | Authors: | Patel, A.B, Radhakrishnan, I, He, Y. | | Deposit date: | 2023-02-22 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of the Saccharomyces cerevisiae Rpd3L histone deacetylase complex.
Nat Commun, 14, 2023
|
|
