3T41
 
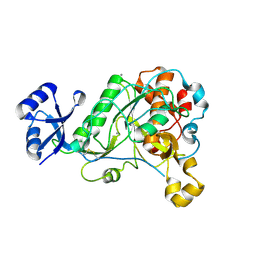 | | 1.95 Angstrom Resolution Crystal Structure of Epidermin Leader Peptide Processing Serine Protease (EpiP) S393A Mutant from Staphylococcus aureus | | Descriptor: | CALCIUM ION, CHLORIDE ION, Epidermin leader peptide processing serine protease EpiP | | Authors: | Minasov, G, Kuhn, M, Ruan, J, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-07-25 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 1.95 Angstrom Resolution Crystal Structure of Epidermin Leader Peptide Processing Serine Protease (EpiP) S393A Mutant from Staphylococcus aureus.
TO BE PUBLISHED
|
|
2L5K
 
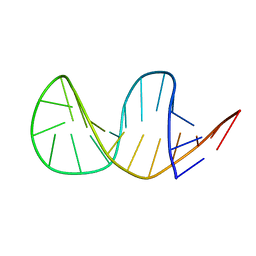 | | Solution structure of truncated 23-mer DNA MUC1 aptamer | | Descriptor: | DNA (5'-R(*(N68)P*G)-D(*CP*AP*GP*TP*TP*GP*AP*TP*CP*CP*TP*TP*TP*GP*GP*AP*TP*AP*CP*CP*CP*TP*GP*GP*T)-3') | | Authors: | Cognet, J, Baouendi, M, Hantz, E, Missailidis, S, Herve du Penhoat, C, Piotto, M. | | Deposit date: | 2010-11-02 | | Release date: | 2011-12-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a truncated anti-MUC1 DNA aptamer determined by mesoscale modeling and NMR.
Febs J., 279, 2012
|
|
6XR4
 
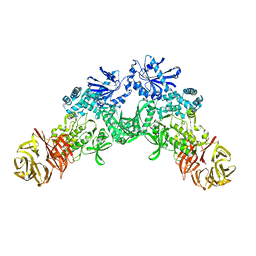 | |
3VK1
 
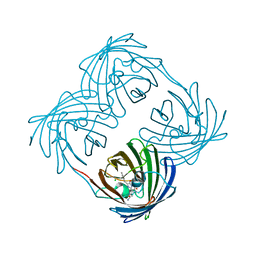 | | Green-fluorescent variant of the non-fluorescent chromoprotein Rtms5 | | Descriptor: | CHLORIDE ION, GFP-like non-fluorescent chromoprotein, IODIDE ION | | Authors: | Battad, J.M, Traore, D.A.K, Wilce, M, Byres, M, Rossjohn, J, Devenish, R.J, Prescott, M. | | Deposit date: | 2011-11-07 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Green Fluorescent Protein Containing a QFG Tri-Peptide Chromophore: Optical Properties and X-Ray Crystal Structure.
Plos One, 7, 2012
|
|
6YB8
 
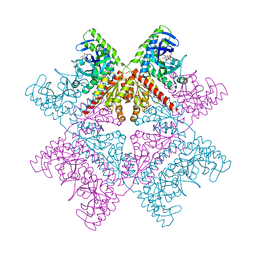 | | Human octameric PAICS in complex with CAIR and SAICAR | | Descriptor: | (2~{S})-2-[[5-azanyl-1-[(2~{R},3~{R},4~{S},5~{R})-3,4-bis(oxidanyl)-5-(phosphonooxymethyl)oxolan-2-yl]imidazol-4-yl]car bonylamino]butanedioic acid, 1,2-ETHANEDIOL, 5-AMINO-1-(5-O-PHOSPHONO-BETA-D-RIBOFURANOSYL)-1H-IMIDAZOLE-4-CARBOXYLIC ACID, ... | | Authors: | Skerlova, J, Unterlass, J, Gottmann, M, Homan, E, Helleday, T, Jemth, A.S, Stenmark, P. | | Deposit date: | 2020-03-16 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Crystal structures of human PAICS reveal substrate and product binding of an emerging cancer target.
J.Biol.Chem., 295, 2020
|
|
6YB9
 
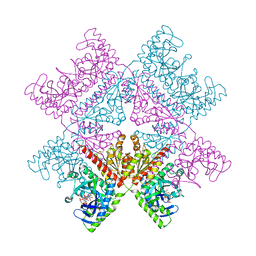 | | Human octameric PAICS in complex with SAICAR, AMP-PNP, and magnesium | | Descriptor: | (2~{S})-2-[[5-azanyl-1-[(2~{R},3~{R},4~{S},5~{R})-3,4-bis(oxidanyl)-5-(phosphonooxymethyl)oxolan-2-yl]imidazol-4-yl]car bonylamino]butanedioic acid, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Skerlova, J, Unterlass, J, Gottmann, M, Homan, E, Helleday, T, Jemth, A.S, Stenmark, P. | | Deposit date: | 2020-03-16 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.406 Å) | | Cite: | Crystal structures of human PAICS reveal substrate and product binding of an emerging cancer target.
J.Biol.Chem., 295, 2020
|
|
6GQU
 
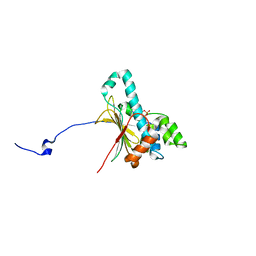 | | Structure of human Heat shock protein 90-alpha N-terminal domain (Hsp90-NTD) variant K112R in complex with cAMP | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Heat shock protein HSP 90-alpha | | Authors: | Tassone, G, Pozzi, C, Mangani, S, Botta, M. | | Deposit date: | 2018-06-08 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Probing the role of Arg97 in Heat shock protein 90 N-terminal domain from the parasite Leishmania braziliensis through site-directed mutagenesis on the human counterpart.
Biochim Biophys Acta Proteins Proteom, 1866, 2018
|
|
6GR4
 
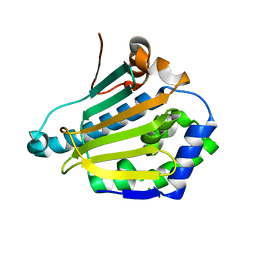 | | Structure of human Heat shock protein 90-alpha N-terminal domain (Hsp90-NTD) variant K112R | | Descriptor: | CHLORIDE ION, Heat shock protein HSP 90-alpha, MAGNESIUM ION | | Authors: | Tassone, G, Pozzi, C, Mangani, S, Botta, M. | | Deposit date: | 2018-06-08 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Probing the role of Arg97 in Heat shock protein 90 N-terminal domain from the parasite Leishmania braziliensis through site-directed mutagenesis on the human counterpart.
Biochim Biophys Acta Proteins Proteom, 1866, 2018
|
|
4A24
 
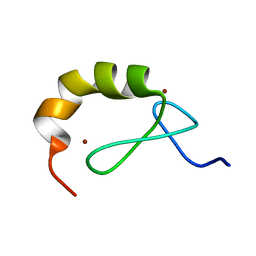 | | Structural and functional analysis of the DEAF-1 and BS69 MYND domains | | Descriptor: | DEFORMED EPIDERMAL AUTOREGULATORY FACTOR 1 HOMOLOG, ZINC ION | | Authors: | Kateb, F, Perrin, H, Tripsianes, K, Zou, P, Spadaccini, R, Bottomley, M, Bepperling, A, Ansieau, S, Sattler, M. | | Deposit date: | 2011-09-22 | | Release date: | 2012-11-07 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Analysis of the Deaf-1 and Bs69 Mynd Domains.
Plos One, 8, 2013
|
|
4AC6
 
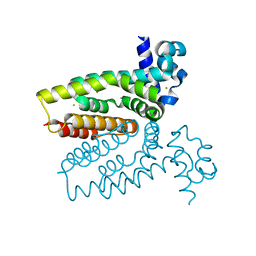 | | Corynebacterium glutamicum AcnR AU derivative structure | | Descriptor: | GOLD ION, HTH-TYPE TRANSCRIPTIONAL REPRESSOR ACNR | | Authors: | Garcia-Nafria, J, Baumgart, M, Turkenburg, J.P, Wilkinson, A.J, Bott, M, Wilson, K.S. | | Deposit date: | 2011-12-14 | | Release date: | 2012-12-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Crystal and Solution Studies Reveal that the Transcriptional Regulator Acnr of Corynebacterium Glutamicum is Regulated by Citrate:Mg2+ Binding to a Non-Canonical Pocket.
J.Biol.Chem., 288, 2013
|
|
1OVD
 
 | | THE K136E MUTANT OF LACTOCOCCUS LACTIS DIHYDROOROTATE DEHYDROGENASE A IN COMPLEX WITH OROTATE | | Descriptor: | DIHYDROOROTATE DEHYDROGENASE A, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Norager, S, Arent, S, Bjornberg, O, Ottosen, M, Lo Leggio, L, Jensen, K.F, Larsen, S. | | Deposit date: | 2003-03-26 | | Release date: | 2003-08-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function.
J.Biol.Chem., 278, 2003
|
|
2YK0
 
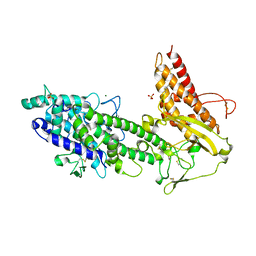 | | Structure of the N-terminal NTS-DBL1-alpha and CIDR-gamma double domain of the PfEMP1 protein from Plasmodium falciparum varO strain. | | Descriptor: | ERYTHROCYTE MEMBRANE PROTEIN 1, MAGNESIUM ION, SULFATE ION | | Authors: | Lewit-Bentley, A, Juillerat, A, Vigan-Womas, I, Guillotte, M, Hessel, A, Raynal, B, Mercereau-Puijalon, O, Bentley, G.A. | | Deposit date: | 2011-05-25 | | Release date: | 2012-05-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for the Abo Blood-Group Dependence of Plasmodium Falciparum Rosetting.
Plos Pathog., 8, 2012
|
|
3VIC
 
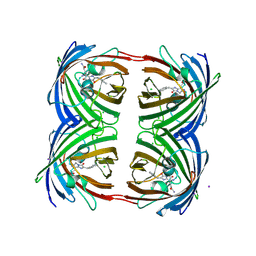 | | Green-fluorescent variant of the non-fluorescent chromoprotein Rtms5 | | Descriptor: | CHLORIDE ION, GFP-like non-fluorescent chromoprotein, IODIDE ION | | Authors: | Battad, J.M, Traore, D.A.K, Byres, E, Wilce, M, Devenish, R.J, Rossjohn, J, Prescott, M. | | Deposit date: | 2011-09-28 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Green Fluorescent Protein Containing a QFG Tri-Peptide Chromophore: Optical Properties and X-Ray Crystal Structure.
Plos One, 7, 2012
|
|
1JQX
 
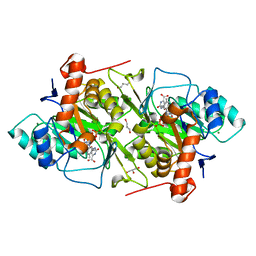 | | The R57A mutant of Lactococcus lactis dihydroorotate dehydrogenase A | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Norager, S, Arent, S, Bjornberg, O, Ottosen, M, Lo Leggio, L, Jensen, K.F, Larsen, S. | | Deposit date: | 2001-08-09 | | Release date: | 2003-09-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function
J.Biol.Chem., 278, 2003
|
|
1JRC
 
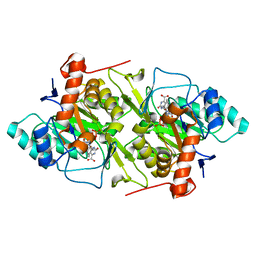 | | The N67A mutant of Lactococcus lactis dihydroorotate dehydrogenase A | | Descriptor: | FLAVIN MONONUCLEOTIDE, OROTIC ACID, dihydroorotate dehydrogenase A | | Authors: | Norager, S, Arent, S, Bjornberg, O, Ottosen, M, Lo Leggio, L, Jensen, K.F, Larsen, S. | | Deposit date: | 2001-08-13 | | Release date: | 2003-09-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function
J.Biol.Chem., 278, 2003
|
|
1JUE
 
 | | 1.8 A resolution structure of native lactococcus lactis dihydroorotate dehydrogenase A | | Descriptor: | ACETIC ACID, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Norager, S, Arent, S, Bjornberg, O, Ottosen, M, Lo Leggio, L, Jensen, K.F, Larsen, S. | | Deposit date: | 2001-08-24 | | Release date: | 2003-09-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function
J.Biol.Chem., 278, 2003
|
|
1PCI
 
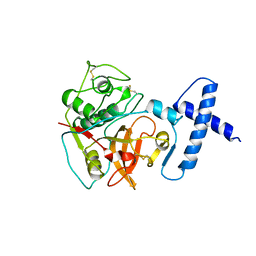 | | PROCARICAIN | | Descriptor: | PROCARICAIN | | Authors: | Groves, M.R, Taylor, M.A.J, Scott, M, Cummings, N.J, Pickersgill, R.W, Jenkins, J.A. | | Deposit date: | 1996-06-28 | | Release date: | 1997-04-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The prosequence of procaricain forms an alpha-helical domain that prevents access to the substrate-binding cleft.
Structure, 4, 1996
|
|
1XQM
 
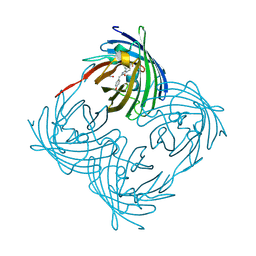 | | Variations on the GFP chromophore scaffold: A fragmented 5-membered heterocycle revealed in the 2.1A crystal structure of a non-fluorescent chromoprotein | | Descriptor: | ACETIC ACID, kindling fluorescent protein | | Authors: | Wilmann, P.G, Petersen, J, Devenish, R.J, Prescott, M, Rossjohn, J. | | Deposit date: | 2004-10-13 | | Release date: | 2004-11-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Variations on the GFP chromophore: A polypeptide fragmentation within the chromophore revealed in the 2.1-A crystal structure of a nonfluorescent chromoprotein from Anemonia sulcata
J.Biol.Chem., 280, 2005
|
|
1UIS
 
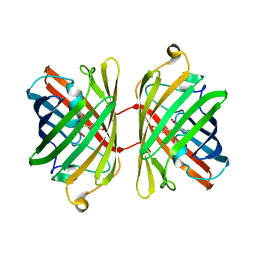 | | The 2.0 crystal structure of eqFP611, a far-red fluorescent protein from the sea anemone Entacmaea quadricolor | | Descriptor: | ACETIC ACID, CALCIUM ION, red fluorescent protein FP611 | | Authors: | Petersen, J, Wilmann, P.G, Beddoe, T, Oakley, A.J, Devenish, R.J, Prescott, M, Rossjohn, J. | | Deposit date: | 2003-07-21 | | Release date: | 2003-10-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2.0A crystal structure of eqFP611, a far-red fluorescent protein from the sea anemone Entacmaea quadricolor
J.Biol.Chem., 278, 2003
|
|
1JQV
 
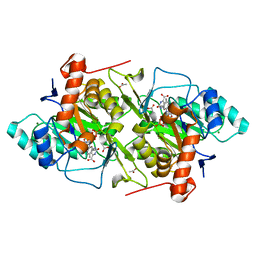 | | The K213E mutant of Lactococcus lactis Dihydroorotate dehydrogenase A | | Descriptor: | ACETIC ACID, Dihydroorotate dehydrogenase A, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Norager, S, Arent, S, Bjornberg, O, Ottosen, M, Lo Leggio, L, Jensen, K.F, Larsen, S. | | Deposit date: | 2001-08-09 | | Release date: | 2003-09-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function
J.Biol.Chem., 278, 2003
|
|
1JRB
 
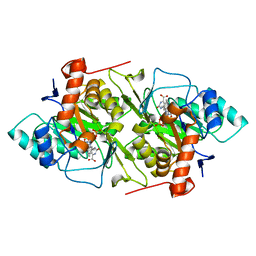 | | The P56A mutant of Lactococcus lactis dihydroorotate dehydrogenase A | | Descriptor: | FLAVIN MONONUCLEOTIDE, OROTIC ACID, dihydroorotate dehydrogenase A | | Authors: | Norager, S, Arent, S, Bjornberg, O, Ottosen, M, Lo Leggio, L, Jensen, K.F, Larsen, S. | | Deposit date: | 2001-08-13 | | Release date: | 2003-09-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function
J.Biol.Chem., 278, 2003
|
|
2P4M
 
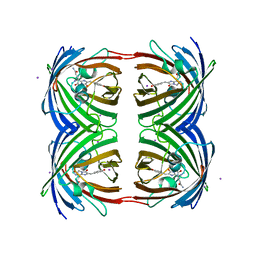 | | High pH structure of Rtms5 H146S variant | | Descriptor: | GFP-like non-fluorescent chromoprotein, IODIDE ION | | Authors: | Battad, J.M, Wilmann, P.G, Olsen, S, Byres, E, Smith, S.C, Dove, S.G, Turcic, K.N, Devenish, R.J, Rossjohn, J, Prescott, M. | | Deposit date: | 2007-03-12 | | Release date: | 2007-04-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A structural basis for the pH-dependent increase in fluorescence efficiency of chromoproteins
J.Mol.Biol., 368, 2007
|
|
5AOQ
 
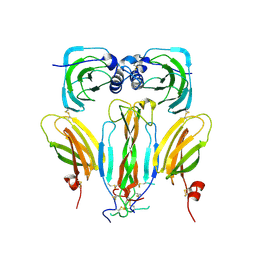 | | Structural basis of neurohormone perception by the receptor tyrosine kinase Torso | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PREPROPTTH, TORSO | | Authors: | Jenni, S, Goyal, Y, von Grotthuss, M, Shvartsman, S.Y, Klein, D.E. | | Deposit date: | 2015-09-11 | | Release date: | 2015-11-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis of Neurohormone Perception by the Receptor Tyrosine Kinase Torso.
Mol.Cell, 60, 2015
|
|
1YZW
 
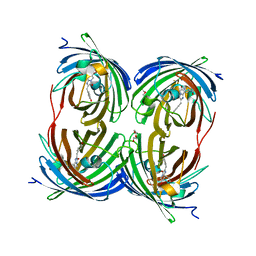 | | The 2.1A Crystal Structure of the Far-red Fluorescent Protein HcRed: Inherent Conformational Flexibility of the Chromophore | | Descriptor: | DI(HYDROXYETHYL)ETHER, GFP-like non-fluorescent chromoprotein | | Authors: | Wilmann, P.G, Petersen, J, Pettikiriarachchi, A, Buckle, A.M, Devenish, R.J, Prescott, M, Rossjohn, J. | | Deposit date: | 2005-02-28 | | Release date: | 2005-05-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The 2.1A Crystal Structure of the Far-red Fluorescent Protein HcRed: Inherent Conformational Flexibility of the Chromophore
J.Mol.Biol., 349, 2005
|
|
1P0Z
 
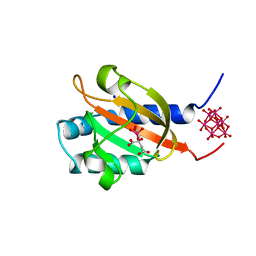 | | Sensor Kinase CitA binding domain | | Descriptor: | CITRATE ANION, MO(VI)(=O)(OH)2 CLUSTER, SODIUM ION, ... | | Authors: | Reinelt, S, Hofmann, E, Gerharz, T, Bott, M, Madden, D.R. | | Deposit date: | 2003-04-11 | | Release date: | 2003-08-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure of the periplasmic ligand-binding domain of the sensor kinase CitA reveals the first extracellular PAS domain.
J.Biol.Chem., 278, 2003
|
|
