8PCN
 
 | | Structure of serine-beta-lactamase CTX-M-14 following the time-resolved active site binding of boric acid and subsequent glycerol-boric acid-ester formation, 100 ms | | Descriptor: | BORATE ION, Beta-lactamase, SULFATE ION, ... | | Authors: | Prester, A, Perbandt, M, Galchenkova, M, Oberthuer, D, Yefanov, O, Hinrichs, W, Rohde, H, Betzel, C. | | Deposit date: | 2023-06-11 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Time-resolved crystallography of boric acid binding to the active site serine of the beta-lactamase CTX-M-14 and subsequent 1,2-diol esterification.
Commun Chem, 7, 2024
|
|
8PCK
 
 | | Structure of serine-beta-lactamase CTX-M-14 following the time-resolved active site binding of boric acid and subsequent glycerol-boric acid-ester formation, 0 ms | | Descriptor: | BORATE ION, Beta-lactamase, SULFATE ION | | Authors: | Prester, A, Perbandt, M, Galchenkova, M, Oberthuer, D, Yefanov, O, Hinrichs, W, Rohde, H, Betzel, C. | | Deposit date: | 2023-06-11 | | Release date: | 2024-06-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Time-resolved crystallography of boric acid binding to the active site serine of the beta-lactamase CTX-M-14 and subsequent 1,2-diol esterification.
Commun Chem, 7, 2024
|
|
8PC9
 
 | | Structure of serine-beta-lactamase CTX-M-14 following the time-resolved active site binding of boric acid, 0 ms - native. | | Descriptor: | Beta-lactamase, SULFATE ION | | Authors: | Prester, A, Perbandt, M, Galchenkova, M, Oberthuer, D, Yefanov, O, Hinrichs, W, Rohde, H, Betzel, C. | | Deposit date: | 2023-06-11 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Time-resolved crystallography of boric acid binding to the active site serine of the beta-lactamase CTX-M-14 and subsequent 1,2-diol esterification.
Commun Chem, 7, 2024
|
|
8PCL
 
 | | Structure of serine-beta-lactamase CTX-M-14 following the time-resolved active site binding of boric acid and subsequent glycerol-boric acid-ester formation, 50 ms | | Descriptor: | BORATE ION, Beta-lactamase, SULFATE ION, ... | | Authors: | Prester, A, Perbandt, M, Galchenkova, M, Oberthuer, D, Yefanov, O, Hinrichs, W, Rohde, H, Betzel, C. | | Deposit date: | 2023-06-11 | | Release date: | 2024-06-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Time-resolved crystallography of boric acid binding to the active site serine of the beta-lactamase CTX-M-14 and subsequent 1,2-diol esterification.
Commun Chem, 7, 2024
|
|
8PCQ
 
 | | Structure of serine-beta-lactamase CTX-M-14 following the time-resolved active site binding of boric acid and subsequent glycerol-boric acid-ester formation, 500 ms | | Descriptor: | BORATE ION, Beta-lactamase, SULFATE ION, ... | | Authors: | Prester, A, Perbandt, M, Galchenkova, M, Oberthuer, D, Yefanov, O, Hinrichs, W, Rohde, H, Betzel, C. | | Deposit date: | 2023-06-11 | | Release date: | 2024-06-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Time-resolved crystallography of boric acid binding to the active site serine of the beta-lactamase CTX-M-14 and subsequent 1,2-diol esterification.
Commun Chem, 7, 2024
|
|
8PCB
 
 | | Structure of serine-beta-lactamase CTX-M-14 following the time-resolved active site binding of boric acid, 80 ms | | Descriptor: | BORATE ION, Beta-lactamase, SULFATE ION | | Authors: | Prester, A, Perbandt, M, Galchenkova, M, Oberthuer, D, Yefanov, O, Hinrichs, W, Rohde, H, Betzel, C. | | Deposit date: | 2023-06-11 | | Release date: | 2024-06-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Time-resolved crystallography of boric acid binding to the active site serine of the beta-lactamase CTX-M-14 and subsequent 1,2-diol esterification.
Commun Chem, 7, 2024
|
|
8PCO
 
 | | Structure of serine-beta-lactamase CTX-M-14 following the time-resolved active site binding of boric acid and subsequent glycerol-boric acid-ester formation, 150 ms | | Descriptor: | BORATE ION, Beta-lactamase, SULFATE ION, ... | | Authors: | Prester, A, Perbandt, M, Galchenkova, M, Oberthuer, D, Yefanov, O, Hinrichs, W, Rohde, H, Betzel, C. | | Deposit date: | 2023-06-11 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Time-resolved crystallography of boric acid binding to the active site serine of the beta-lactamase CTX-M-14 and subsequent 1,2-diol esterification.
Commun Chem, 7, 2024
|
|
8PCC
 
 | | Structure of serine-beta-lactamase CTX-M-14 following the time-resolved active site binding of boric acid, 100 ms | | Descriptor: | BORATE ION, Beta-lactamase, SULFATE ION | | Authors: | Prester, A, Perbandt, M, Galchenkova, M, Oberthuer, D, Yefanov, O, Hinrichs, W, Rohde, H, Betzel, C. | | Deposit date: | 2023-06-11 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Time-resolved crystallography of boric acid binding to the active site serine of the beta-lactamase CTX-M-14 and subsequent 1,2-diol esterification.
Commun Chem, 7, 2024
|
|
8PCV
 
 | | Structure of serine-beta-lactamase CTX-M-14 following the time-resolved active site binding of boric acid and subsequent glycerol-boric acid-ester formation, 10000 ms | | Descriptor: | BORATE ION, Beta-lactamase, SULFATE ION, ... | | Authors: | Prester, A, Perbandt, M, Galchenkova, M, Oberthuer, D, Yefanov, O, Hinrichs, W, Rohde, H, Betzel, C. | | Deposit date: | 2023-06-11 | | Release date: | 2024-06-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Time-resolved crystallography of boric acid binding to the active site serine of the beta-lactamase CTX-M-14 and subsequent 1,2-diol esterification.
Commun Chem, 7, 2024
|
|
5U5Q
 
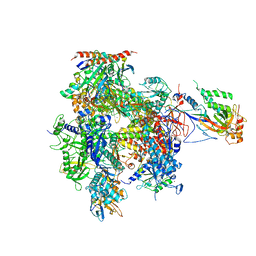 | | 12 Subunit RNA Polymerase II at Room Temperature collected using SFX | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Bushnell, D.A, Oberthur, D, Mariani, V, Yefanov, O, Tolstikova, A, Barty, A. | | Deposit date: | 2016-12-07 | | Release date: | 2017-03-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Double-flow focused liquid injector for efficient serial femtosecond crystallography.
Sci Rep, 7, 2017
|
|
4R8I
 
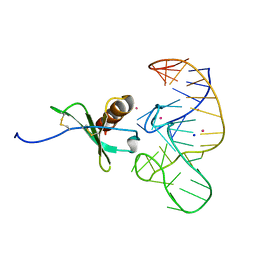 | | High Resolution Structure of a Mirror-Image RNA Oligonucleotide Aptamer in Complex with the Chemokine CCL2 | | Descriptor: | C-C motif chemokine 2, Mirror-Image RNA Oligonucleotide Aptamer NOXE36, POTASSIUM ION, ... | | Authors: | Oberthuer, D, Achenbach, J, Gabdulkhakov, A, Falke, S, Buchner, K, Maasch, C, Rehders, D, Klussmann, S, Betzel, C. | | Deposit date: | 2014-09-02 | | Release date: | 2015-04-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of a mirror-image L-RNA aptamer (Spiegelmer) in complex with the natural L-protein target CCL2.
Nat Commun, 6, 2015
|
|
2XSL
 
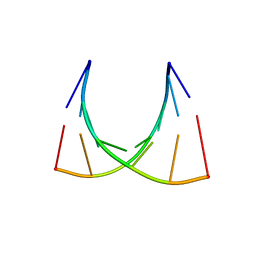 | | The crystal structure of a Thermus thermophilus tRNAGly acceptor stem microhelix at 1.6 Angstroem resolution | | Descriptor: | 5'-R(*CP*UP*CP*CP*CP*GP*C)-3', 5'-R(*GP*CP*GP*GP*GP*AP*G)-3' | | Authors: | Oberthuer, D, Eichert, A, Erdmann, V.A, Fuerste, J.P, Betzel, C, Foerster, C. | | Deposit date: | 2010-10-29 | | Release date: | 2011-08-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | The Crystal Structure of a Thermus Thermophilus tRNA(Gly) Acceptor Stem Microhelix at 1.6 A Resolution.
Biochem.Biophys.Res.Commun., 404, 2011
|
|
4WLA
 
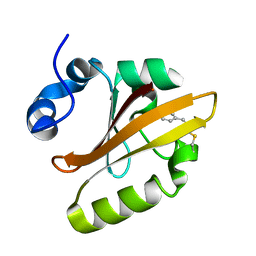 | |
8S8W
 
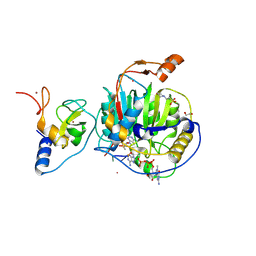 | | SARS-CoV-2 nsp10-16 methyltransferase in complex with Sangivamycin and m7GpppA-RNA (Cap0-RNA) | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D, Scheer, T.E.S. | | Deposit date: | 2024-03-07 | | Release date: | 2024-03-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | SARS-CoV-2 methyltransferase nsp10-16 in complex with natural and drug-like purine analogs for guiding structure-based drug discovery
Elife, 2024
|
|
8S8X
 
 | | SARS-CoV-2 nsp10-16 methyltransferase in complex with Toyocamycin and m7GpppA-RNA (Cap0-RNA) | | Descriptor: | 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-amino-7-(beta-D-ribofuranosyl)-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D, Scheer, T.E.S. | | Deposit date: | 2024-03-07 | | Release date: | 2024-03-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | SARS-CoV-2 methyltransferase nsp10-16 in complex with natural and drug-like purine analogs for guiding structure-based drug discovery
Elife, 2024
|
|
4WL9
 
 | |
4X32
 
 | | Bacteriorhodopsin ground state structure collected in cryo conditions from crystals obtained in LCP with PEG as a precipitant. | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin, RETINAL | | Authors: | Nogly, P, Standfuss, J. | | Deposit date: | 2014-11-27 | | Release date: | 2015-02-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Lipidic cubic phase serial millisecond crystallography using synchrotron radiation.
Iucrj, 2, 2015
|
|
9EMV
 
 | | SARS-CoV-2 nsp10-16 methyltransferase in complex with Sangivamycin and m7GpppA (Cap0-analog)/m7GpppAm (Cap1-analog) | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D, Scheer, T.E.S. | | Deposit date: | 2024-03-11 | | Release date: | 2024-03-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | SARS-CoV-2 methyltransferase nsp10-16 in complex with natural and drug-like purine analogs for guiding structure-based drug discovery
Elife, 2024
|
|
9EML
 
 | | SARS-CoV-2 methyltransferase nsp10-16 in complex with SAM and m7GpppA (Cap0-analog)/m7GpppAm (Cap1-analog) | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D, Kiene, A. | | Deposit date: | 2024-03-08 | | Release date: | 2024-03-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | SARS-CoV-2 methyltransferase nsp10-16 in complex with natural and drug-like purine analogs for guiding structure-based drug discovery
Elife, 2024
|
|
8BSD
 
 | | SARS-CoV-2 nsp10-16 methyltransferase in complex with tubercidin | | Descriptor: | '2-(4-AMINO-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, ... | | Authors: | Kremling, V, Oberthuer, D, Sprenger, J. | | Deposit date: | 2022-11-24 | | Release date: | 2022-12-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | SARS-CoV-2 methyltransferase nsp10-16 in complex with natural and drug-like purine analogs for guiding structure-based drug discovery
Elife, 2024
|
|
8BZV
 
 | | SARS-CoV-2 nsp10-16 methyltransferase in complex with adenosine | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D. | | Deposit date: | 2022-12-15 | | Release date: | 2023-01-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | SARS-CoV-2 methyltransferase nsp10-16 in complex with natural and drug-like purine analogs for guiding structure-based drug discovery
Elife, 2024
|
|
8C5M
 
 | | SARS-CoV-2 nsp10-16 methyltransferase in complex with MTA | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D, Falke, S. | | Deposit date: | 2023-01-09 | | Release date: | 2023-03-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | SARS-CoV-2 methyltransferase nsp10-16 in complex with natural and drug-like purine analogs for guiding structure-based drug discovery
Elife, 2024
|
|
5G0Z
 
 | | Structure of native granulovirus polyhedrin determined using an X-ray free-electron laser | | Descriptor: | GRANULIN | | Authors: | Gati, C, Bunker, R.D, Oberthur, D, Metcalf, P, Henry, C. | | Deposit date: | 2016-03-23 | | Release date: | 2017-02-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Atomic structure of granulin determined from native nanocrystalline granulovirus using an X-ray free-electron laser.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5G3X
 
 | | Structure of recombinant granulovirus polyhedrin | | Descriptor: | GRANULOVIRUS POLYHEDRIN | | Authors: | Bunker, R.D, Chiu, E, Metcalf, P. | | Deposit date: | 2016-05-02 | | Release date: | 2017-02-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Atomic structure of granulin determined from native nanocrystalline granulovirus using an X-ray free-electron laser.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8RTG
 
 | |
