2J40
 
 | | 1-pyrroline-5-carboxylate dehydrogenase from Thermus thermophilus with bound inhibitor L-proline and NAD. | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 1-PYRROLINE-5-CARBOXYLATE DEHYDROGENASE, ... | | Authors: | Inagaki, E, Sakamoto, K, Nishio, M, Yokoyama, S. | | Deposit date: | 2006-08-24 | | Release date: | 2007-10-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Ternary Complex of Delta1-Pyrroline-5-Carboxylate Dehydrogenase with Substrate Mimic and Co-Factoer
To be Published
|
|
1WNS
 
 | | Crystal structure of family B DNA polymerase from hyperthermophilic archaeon pyrococcus kodakaraensis KOD1 | | Descriptor: | DNA POLYMERASE | | Authors: | Hashimoto, H, Inoue, T, Kai, Y, Fujiwara, S, Takagi, M, Nishioka, M, Imanaka, T. | | Deposit date: | 2004-08-09 | | Release date: | 2004-08-17 | | Last modified: | 2017-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of DNA Polymerase from Hyperthermophilic Archaeon Pyrococcus Kodakaraensis Kod1
J.Mol.Biol., 306, 2001
|
|
3BUZ
 
 | | Crystal structure of ia-bTAD-actin complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Tsuge, H, Nagahama, M, Oda, M, Iwamoto, S, Utsunomiya, H, Marquez, V.E, Katunuma, N, Nishizawa, M, Sakurai, J. | | Deposit date: | 2008-01-04 | | Release date: | 2008-05-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structural basis of actin recognition and arginine ADP-ribosylation by Clostridium perfringens iota-toxin
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
5X3D
 
 | | Crystal structure of HEP-CMP-bound form of cytidylyltransferase (CyTase) domain of Fom1 from Streptomyces wedmorensis | | Descriptor: | Phosphoenolpyruvate phosphomutase, [[(2R,3S,4R,5R)-5-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-(2-hydroxyethyl)phosphinic acid | | Authors: | Tomita, T, Cho, S.H, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2017-02-04 | | Release date: | 2017-09-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Fosfomycin Biosynthesis via Transient Cytidylylation of 2-Hydroxyethylphosphonate by the Bifunctional Fom1 Enzyme
ACS Chem. Biol., 12, 2017
|
|
5XOY
 
 | | Crystal structure of LysK from Thermus thermophilus in complex with Lysine | | Descriptor: | LYSINE, SULFATE ION, [LysW]-lysine hydrolase | | Authors: | Tomita, T, Fujita, S, Hasebe, F, Cho, S.-H, Yoshida, A, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2017-05-31 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystal structure of LysK, an enzyme catalyzing the last step of lysine biosynthesis in Thermus thermophilus, in complex with lysine: Insight into the mechanism for recognition of the amino-group carrier protein, LysW
Biochem. Biophys. Res. Commun., 491, 2017
|
|
3WFZ
 
 | | Crystal structure of Galacto-N-Biose/Lacto-N-Biose I Phosphorylase C236Y Mutant | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, Lacto-N-biose phosphorylase | | Authors: | Koyama, Y, Hidaka, M, Kawakami, M, Nishimoto, M, Kitaoka, M. | | Deposit date: | 2013-07-25 | | Release date: | 2013-10-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Directed evolution to enhance thermostability of galacto-N-biose/lacto-N-biose I phosphorylase.
Protein Eng.Des.Sel., 26, 2013
|
|
5B1Q
 
 | | Human herpesvirus 6B tegument protein U14 | | Descriptor: | GLYCEROL, U14 protein | | Authors: | Wang, B, Nishimura, M, Tang, H, Kawabata, A, Mahmoud, N.F, Khanlari, Z, Hamada, D, Tsuruta, H, Mori, Y. | | Deposit date: | 2015-12-11 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Human Herpesvirus 6B Tegument Protein U14.
Plos Pathog., 12, 2016
|
|
3VPB
 
 | | ArgX from Sulfolobus tokodaii complexed with LysW/Glu/ADP/Mg/Zn/Sulfate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Alpha-aminoadipate carrier protein lysW, GLUTAMIC ACID, ... | | Authors: | Tomita, T, Ouchi, T, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2012-02-29 | | Release date: | 2013-02-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Lysine and arginine biosyntheses mediated by a common carrier protein in Sulfolobus
Nat.Chem.Biol., 9, 2013
|
|
3AAW
 
 | | Crystal structure of aspartate kinase from Corynebacterium glutamicum in complex with lysine and threonine | | Descriptor: | Aspartokinase, Aspartokinase LysC beta subunit, LYSINE, ... | | Authors: | Yoshida, A, Tomita, T, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2009-11-27 | | Release date: | 2010-06-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism of concerted inhibition of {alpha}2{beta}2-type heterooligomeric aspartate kinase from Corynebacterium glutamicum
J.Biol.Chem., 285, 2010
|
|
3AB2
 
 | | Crystal structure of aspartate kinase from Corynebacterium glutamicum in complex with threonine | | Descriptor: | Aspartokinase, THREONINE | | Authors: | Yoshida, A, Tomita, T, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2009-11-30 | | Release date: | 2010-06-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Mechanism of concerted inhibition of {alpha}2{beta}2-type heterooligomeric aspartate kinase from Corynebacterium glutamicum
J.Biol.Chem., 285, 2010
|
|
3VPD
 
 | | LysX from Thermus thermophilus complexed with AMP-PNP | | Descriptor: | CITRIC ACID, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Ribosomal protein S6 modification protein, ... | | Authors: | Tomita, T, Ouchi, T, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2012-02-29 | | Release date: | 2013-02-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Lysine and arginine biosyntheses mediated by a common carrier protein in Sulfolobus.
Nat.Chem.Biol., 9, 2013
|
|
1JQE
 
 | | Crystal Structure Analysis of Human Histamine Methyltransferase (Ile105 Polymorphic Variant) Complexed with AdoHcy and Antimalarial Drug Quinacrine | | Descriptor: | Histamine N-Methyltransferase, QUINACRINE, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Horton, J.R, Sawada, K, Nishibori, M, Zhang, X, Cheng, X. | | Deposit date: | 2001-08-06 | | Release date: | 2002-08-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Two polymorphic forms of human histamine methyltransferase: structural, thermal, and kinetic comparisons.
Structure, 9, 2001
|
|
2Z1Y
 
 | | Crystal structure of LysN, alpha-aminoadipate aminotransferase (complexed with N-(5'-phosphopyridoxyl)-L-leucine), from Thermus thermophilus HB27 | | Descriptor: | Alpha-aminodipate aminotransferase, LEUCINE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Tomita, T, Miyazaki, T, Miyagawa, T, Fushinobu, S, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2007-05-16 | | Release date: | 2008-06-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Mechanism of broad substrate specificity of alpha-aminoadipate aminotransferase from Thermus thermophilus
To be Published
|
|
3CBF
 
 | | Crystal structure of LysN, alpha-aminoadipate aminotransferase, from Thermus thermophilus HB27 | | Descriptor: | (2S)-2-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]hexanedioic acid, Alpha-aminodipate aminotransferase | | Authors: | Tomita, T, Miyazaki, T, Miyagawa, T, Fushinobu, S, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2008-02-21 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Mechanism for multiple-substrates recognition of alpha-aminoadipate aminotransferase from Thermus thermophilus
Proteins, 2008
|
|
1JQD
 
 | | Crystal Structure Analysis of Human Histamine Methyltransferase (Thr105 Polymorphic Variant) Complexed with AdoHcy and Histamine | | Descriptor: | HISTAMINE, Histamine N-Methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Horton, J.R, Sawada, K, Nishibori, M, Zhang, X, Cheng, X. | | Deposit date: | 2001-08-06 | | Release date: | 2002-08-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Two polymorphic forms of human histamine methyltransferase: structural, thermal, and kinetic comparisons.
Structure, 9, 2001
|
|
2ZP7
 
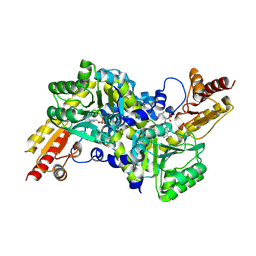 | | Crystal structure of LysN, alpha-aminoadipate aminotransferase (Leucine complex), from Thermus thermophilus HB27 | | Descriptor: | Alpha-aminodipate aminotransferase, LEUCINE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Tomita, T, Miyazaki, T, Miyagawa, T, Fushinobu, S, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2008-06-30 | | Release date: | 2009-01-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Mechanism for multiple-substrates recognition of alpha-aminoadipate aminotransferase from Thermus thermophilus
Proteins, 2008
|
|
3VPC
 
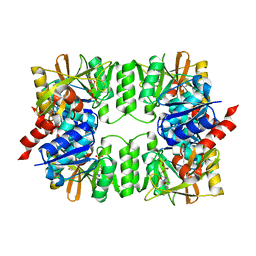 | | ArgX from Sulfolobus tokodaii complexed with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Putative acetylornithine deacetylase | | Authors: | Tomita, T, Horie, A, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2012-02-29 | | Release date: | 2013-02-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Lysine and arginine biosyntheses mediated by a common carrier protein in Sulfolobus.
Nat.Chem.Biol., 9, 2013
|
|
3AB4
 
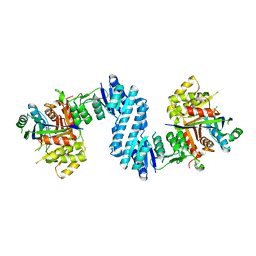 | | Crystal structure of feedback inhibition resistant mutant of aspartate kinase from Corynebacterium glutamicum in complex with lysine and threonine | | Descriptor: | Aspartokinase, LYSINE, THREONINE | | Authors: | Yoshida, A, Tomita, T, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2009-11-30 | | Release date: | 2010-06-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Mechanism of concerted inhibition of {alpha}2{beta}2-type heterooligomeric aspartate kinase from Corynebacterium glutamicum
J.Biol.Chem., 285, 2010
|
|
6ITD
 
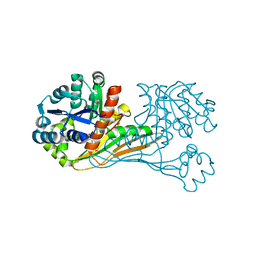 | | Crystal structure of BioU (K124A) from Synechocystis sp.PCC6803 in complex with the analog of reaction intermediate, 3-(1-aminoethyl)-nonanedioic acid | | Descriptor: | 3-(1-AMINOETHYL)NONANEDIOIC ACID, Slr0355 protein | | Authors: | Sakaki, K, Tomita, T, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2018-11-21 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A suicide enzyme catalyzes multiple reactions for biotin biosynthesis in cyanobacteria.
Nat.Chem.Biol., 16, 2020
|
|
7WUX
 
 | | Crystal structure of AziU3/U2 complexed with (5S,6S)-O7-sulfo DADH from Streptomyces sahachiroi | | Descriptor: | (2S,5S,6S)-2,6-bis(azanyl)-5-oxidanyl-7-sulfooxy-heptanoic acid, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, AziU2, ... | | Authors: | Kurosawa, S, Yoshida, A, Tomita, T, Nishiyama, M. | | Deposit date: | 2022-02-09 | | Release date: | 2022-09-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Basis for Enzymatic Aziridine Formation via Sulfate Elimination.
J.Am.Chem.Soc., 144, 2022
|
|
7WUW
 
 | | Crystal structure of AziU3/U2 from Streptomyces sahachiroi | | Descriptor: | AziU2, AziU3, MAGNESIUM ION, ... | | Authors: | Kurosawa, S, Yoshida, A, Tomita, T, Nishiyama, M. | | Deposit date: | 2022-02-09 | | Release date: | 2022-09-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Molecular Basis for Enzymatic Aziridine Formation via Sulfate Elimination.
J.Am.Chem.Soc., 144, 2022
|
|
6IR4
 
 | | Crystal structure of BioU from Synechocystis sp.PCC6803 (apo form) | | Descriptor: | Slr0355 protein | | Authors: | Sakaki, K, Oishi, K, Shimizu, T, Tomita, T, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2018-11-10 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A suicide enzyme catalyzes multiple reactions for biotin biosynthesis in cyanobacteria.
Nat.Chem.Biol., 16, 2020
|
|
6K36
 
 | |
1V8B
 
 | | Crystal structure of a hydrolase | | Descriptor: | ADENOSINE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, adenosylhomocysteinase | | Authors: | Tanaka, N, Nakanishi, M, Kusakabe, Y, Shiraiwa, K, Kitade, Y, Nakamura, K.T. | | Deposit date: | 2004-01-03 | | Release date: | 2004-10-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of S-Adenosyl-l-Homocysteine Hydrolase from the Human Malaria Parasite Plasmodium falciparum
J.Mol.Biol., 343, 2004
|
|
7Y5Y
 
 | | X-ray Structure of Stay-Green (SGR) from Anaerolineae bacterium. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PHOSPHATE ION, ... | | Authors: | Dey, D, Nishijima, M, Kurisu, G, Tanaka, H, Ito, H. | | Deposit date: | 2022-06-18 | | Release date: | 2022-10-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure and reaction mechanism of a bacterial Mg-dechelatase homolog from the Chloroflexi Anaerolineae.
Protein Sci., 31, 2022
|
|
