4V1T
 
 | |
4V1V
 
 | | Heterocyclase in complex with substrate and Cofactor | | Descriptor: | LYND, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Koehnke, J, Naismith, J.H. | | Deposit date: | 2014-10-02 | | Release date: | 2015-01-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural Analysis of Leader Peptide Binding Enables Leader-Free Cyanobactin Processing.
Nat.Chem.Biol., 11, 2015
|
|
4UW0
 
 | |
5JQG
 
 | | An apo tubulin-RB-TTL complex structure used for side-by-side comparison | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wang, Y.X, Naismith, J.H, Zhu, X. | | Deposit date: | 2016-05-04 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Pironetin reacts covalently with cysteine-316 of alpha-tubulin to destabilize microtubule
Nat Commun, 7, 2016
|
|
8OWV
 
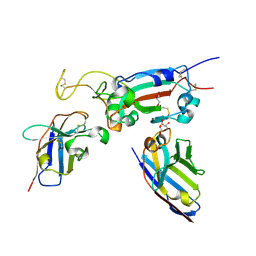 | | H6 and F2 nanobodies bound to SARS-CoV-2 spike RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, F2, GLYCEROL, ... | | Authors: | Mikolajek, H, Naismith, J.H, Owens, R.J. | | Deposit date: | 2023-04-28 | | Release date: | 2024-05-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural and functional characterization of nanobodies that neutralize Omicron variants of SARS-CoV-2.
Open Biology, 14, 2024
|
|
8OYU
 
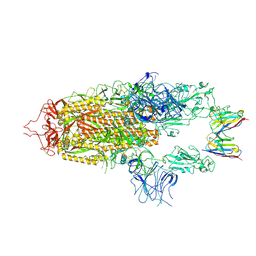 | | Stabilised BA.1 SARS-CoV-2 spike with H6 nanobodies in '2 up 1 down' RBD conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, H6 nanobody, ... | | Authors: | Weckener, M, Naismith, J.H, Owens, R.J. | | Deposit date: | 2023-05-05 | | Release date: | 2024-05-15 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural and functional characterization of nanobodies that neutralize Omicron variants of SARS-CoV-2.
Open Biology, 14, 2024
|
|
8OYT
 
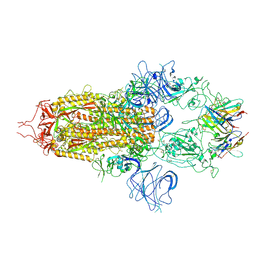 | | Stabilised BA.1 SARS-CoV-2 spike with H6 nanobodies in '3 up' RBD conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, H6 nanobody, ... | | Authors: | Weckener, M, Naismith, J.H, Owens, R.J. | | Deposit date: | 2023-05-05 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural and functional characterization of nanobodies that neutralize Omicron variants of SARS-CoV-2.
Open Biology, 14, 2024
|
|
8OWT
 
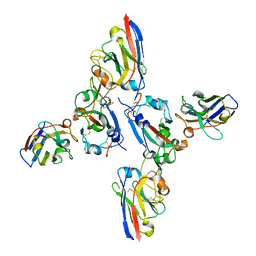 | | SARS-CoV-2 spike RBD with A8 and H3 nanobodies bound | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody A8, ... | | Authors: | Mikolajek, H, Naismith, J.H, Owens, R.J. | | Deposit date: | 2023-04-28 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural and functional characterization of nanobodies that neutralize Omicron variants of SARS-CoV-2.
Open Biology, 14, 2024
|
|
8OWW
 
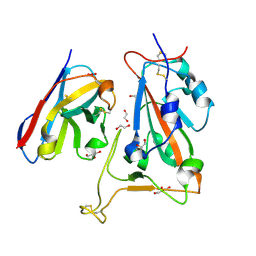 | | B5-5 nanobody bound to SARS-CoV-2 spike RBD (Wuhan) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, B5-5 nanobody, ... | | Authors: | Cornish, K.A.S, Naismith, J.H, Owens, R.J. | | Deposit date: | 2023-04-28 | | Release date: | 2024-05-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.969 Å) | | Cite: | Structural and functional characterization of nanobodies that neutralize Omicron variants of SARS-CoV-2.
Open Biology, 14, 2024
|
|
5LDT
 
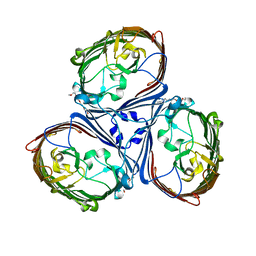 | | Crystal Structures of MOMP from Campylobacter jejuni | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, CALCIUM ION, MOMP porin | | Authors: | Ferrara, L.G.M, Wallat, G.D, Moynie, L, Naismith, J.H. | | Deposit date: | 2016-06-27 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | MOMP from Campylobacter jejuni Is a Trimer of 18-Stranded beta-Barrel Monomers with a Ca(2+) Ion Bound at the Constriction Zone.
J.Mol.Biol., 428, 2016
|
|
5LQ4
 
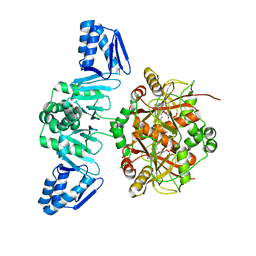 | | The Structure of ThcOx, the First Oxidase Protein from the Cyanobactin Pathways | | Descriptor: | CyaGox, FLAVIN MONONUCLEOTIDE | | Authors: | Bent, A.F, Wagner, A, Naismith, J.H. | | Deposit date: | 2016-08-16 | | Release date: | 2016-11-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of the cyanobactin oxidase ThcOx from Cyanothece sp. PCC 7425, the first structure to be solved at Diamond Light Source beamline I23 by means of S-SAD.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5LMZ
 
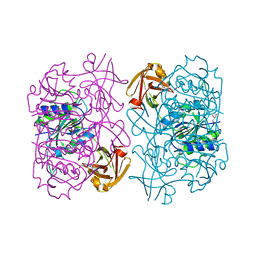 | | Fluorinase from Streptomyces sp. MA37 | | Descriptor: | 1-DEAZA-ADENOSINE, CHLORIDE ION, Fluorinase, ... | | Authors: | Mann, G, O'Hagan, D, Naismith, J.H, Bandaranayaka, N. | | Deposit date: | 2016-08-02 | | Release date: | 2016-08-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Identification of fluorinases from Streptomyces sp MA37, Norcardia brasiliensis, and Actinoplanes sp N902-109 by genome mining.
Chembiochem, 15, 2014
|
|
2IYD
 
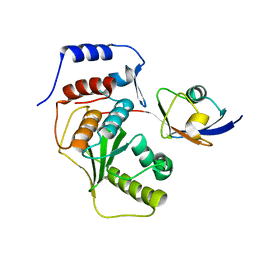 | | SENP1 covalent complex with SUMO-2 | | Descriptor: | SENTRIN-SPECIFIC PROTEASE 1, SMALL UBIQUITIN-RELATED MODIFIER 2 | | Authors: | Dong, C, Naismith, J.H. | | Deposit date: | 2006-07-14 | | Release date: | 2006-08-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Senp1 Native Structure
To be Published
|
|
2IYC
 
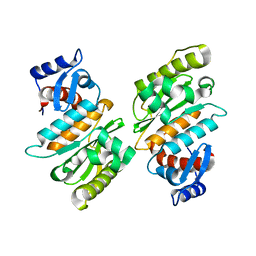 | | SENP1 native structure | | Descriptor: | SENTRIN-SPECIFIC PROTEASE 1 | | Authors: | Dong, C, Naismith, J.H. | | Deposit date: | 2006-07-14 | | Release date: | 2006-08-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Senp1 Native Structure
To be Published
|
|
6Q5E
 
 | | Crystal structure of the ferric enterobactin receptor from Pseudomonas aeruginosa (PfeA) in complex with enterobactin | | Descriptor: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, FE (III) ION, Ferric enterobactin receptor, ... | | Authors: | Moynie, L, Naismith, J.H. | | Deposit date: | 2018-12-07 | | Release date: | 2019-01-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The complex of ferric-enterobactin with its transporter from Pseudomonas aeruginosa suggests a two-site model.
Nat Commun, 10, 2019
|
|
6QGM
 
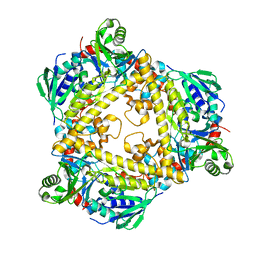 | | VirX1 apo structure | | Descriptor: | VirX1 | | Authors: | Gkotsi, D.S, Ludewig, H, Sharma, S.V, Unsworth, W.P, Taylor, R.J.K, McLachlan, M.M.W, Shanahan, S, Naismith, J.H, Goss, R.J.M. | | Deposit date: | 2019-01-11 | | Release date: | 2019-10-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A marine viral halogenase that iodinates diverse substrates.
Nat.Chem., 11, 2019
|
|
2JKC
 
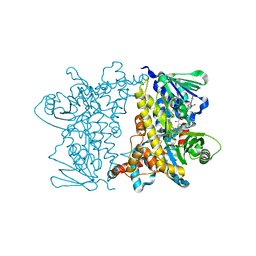 | | Crystal Structure of E346D of Tryptophan 7-Halogenase (PrnA) | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Flavin-dependent tryptophan halogenase PrnA, ... | | Authors: | Zhu, X, Naismith, J.H. | | Deposit date: | 2008-08-26 | | Release date: | 2008-09-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | New insights into the mechanism of enzymatic chlorination of tryptophan.
Angew. Chem. Int. Ed. Engl., 47, 2008
|
|
4FQ9
 
 | | Crystal Structure of 3-hydroxydecanoyl-Acyl Carrier Protein Dehydratase (FabA) from Pseudomonas aeruginosa | | Descriptor: | 3-hydroxydecanoyl-[acyl-carrier-protein] dehydratase, GLYCEROL, PHOSPHATE ION | | Authors: | Moynie, L, Mcmahon, S.A, Duthie, F.G, Naismith, J.H. | | Deposit date: | 2012-06-25 | | Release date: | 2013-03-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural insights into the mechanism and inhibition of the beta-hydroxydecanoyl-acyl carrier protein dehydratase from Pseudomonas aeruginosa
J.Mol.Biol., 425, 2013
|
|
7NHR
 
 | |
7OBW
 
 | | Crystal structure of the ferric enterobactin receptor (PfeA) from Pseudomonas aeruginosa in complex with TCV-L6 | | Descriptor: | FE (III) ION, Ferric enterobactin receptor, [4-[[[(2~{S})-2-[[2,3-bis(oxidanyl)phenyl]carbonylamino]-3-[[(2~{S})-2-[[2,3-bis(oxidanyl)phenyl]carbonylamino]-3-[2-[[2,3-bis(oxidanyl)phenyl]carbonylamino]ethanoylamino]propanoyl]amino]propanoyl]amino]methyl]-1,2,3-triazol-1-yl]methyl 4-[4-[(5~{S})-5-(acetamidomethyl)-2-oxidanylidene-1,3-oxazolidin-3-yl]-2-fluoranyl-phenyl]piperazine-1-carboxylate | | Authors: | Moynie, L, Naismith, J.H. | | Deposit date: | 2021-04-23 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Hijacking of the Enterobactin Pathway by a Synthetic Catechol Vector Designed for Oxazolidinone Antibiotic Delivery in Pseudomonas aeruginosa .
Acs Infect Dis., 8, 2022
|
|
6FCA
 
 | | Catalytic subunit HisG from Psychrobacter arcticus ATP phosphoribosyltransferase (HisZG ATPPRT) in complex with PRPP | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, ATP phosphoribosyltransferase | | Authors: | Alphey, M.S, Ge, Y, Fisher, G, Czekster, C.M, Naismith, J.H, da Silva, R.G. | | Deposit date: | 2017-12-20 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Catalytic and Anticatalytic Snapshots of a Short-Form ATP Phosphoribosyltransferase
Acs Catalysis, 2018
|
|
6FCC
 
 | | Catalytic subunit HisG from Psychrobacter arcticus ATP phosphoribosyltransferase (HisZG ATPPRT) | | Descriptor: | ATP phosphoribosyltransferase, L(+)-TARTARIC ACID | | Authors: | Alphey, M.S, Ge, Y, Fisher, G, Czekster, C.M, Naismith, J.H, da Silva, R.G. | | Deposit date: | 2017-12-20 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Catalytic and Anticatalytic Snapshots of a Short-Form ATP Phosphoribosyltransferase
Acs Catalysis, 2018
|
|
6FCW
 
 | | Catalytic subunit HisG from Psychrobacter arcticus ATP phosphoribosyltransferase (HisZG ATPPRT) in complex with PRATP | | Descriptor: | ATP phosphoribosyltransferase, MAGNESIUM ION, PHOSPHORIBOSYL ATP | | Authors: | Alphey, M.S, Ge, Y, Fisher, G, Czekster, C.M, Naismith, J.H, da Silva, R.G. | | Deposit date: | 2017-12-21 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Catalytic and Anticatalytic Snapshots of a Short-Form ATP Phosphoribosyltransferase
Acs Catalysis, 2018
|
|
6FCT
 
 | | Catalytic subunit HisG from Psychrobacter arcticus ATP phosphoribosyltransferase (HisZG ATPPRT) in complex with PRPP and ATP | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, ADENOSINE-5'-TRIPHOSPHATE, ATP phosphoribosyltransferase, ... | | Authors: | Alphey, M.S, Ge, Y, Fisher, G, Czekster, C.M, Naismith, J.H, da Silva, R.G. | | Deposit date: | 2017-12-21 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Catalytic and Anticatalytic Snapshots of a Short-Form ATP Phosphoribosyltransferase
Acs Catalysis, 2018
|
|
6FCY
 
 | | Catalytic subunit HisG from Psychrobacter arcticus ATP phosphoribosyltransferase (HisZG ATPPRT) in complex with PRPP and ADP | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, ADENOSINE-5'-DIPHOSPHATE, ATP phosphoribosyltransferase, ... | | Authors: | Alphey, M.S, Ge, Y, Fisher, G, Czekster, C.M, Naismith, J.H, da Silva, R.G. | | Deposit date: | 2017-12-21 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Catalytic and Anticatalytic Snapshots of a Short-Form ATP Phosphoribosyltransferase
Acs Catalysis, 2018
|
|
