5TR1
 
 | |
6UQF
 
 | | Human HCN1 channel in a hyperpolarized conformation | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, MERCURY (II) ION, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 | | Authors: | Lee, C.-H, MacKinnon, R. | | Deposit date: | 2019-10-19 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Voltage Sensor Movements during Hyperpolarization in the HCN Channel.
Cell, 179, 2019
|
|
5VA3
 
 | |
5VA1
 
 | |
5U6O
 
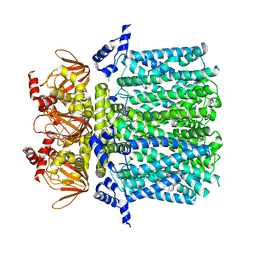 | |
6UWM
 
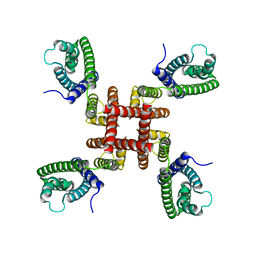 | | Single particle cryo-EM structure of KvAP | | Descriptor: | Voltage-gated potassium channel | | Authors: | Tao, X, MacKinnon, R. | | Deposit date: | 2019-11-05 | | Release date: | 2019-12-04 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Cryo-EM structure of the KvAP channel reveals a non-domain-swapped voltage sensor topology.
Elife, 8, 2019
|
|
5U76
 
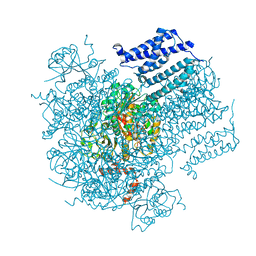 | |
6V3G
 
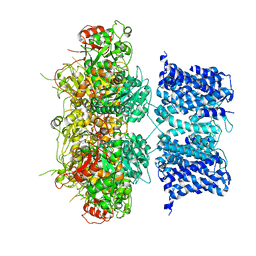 | | Cryo-EM structure of Ca2+-free hsSlo1 channel | | Descriptor: | Calcium-activated potassium channel subunit alpha-1 | | Authors: | Tao, X, MacKinnon, R. | | Deposit date: | 2019-11-25 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Molecular structures of the human Slo1 K + channel in complex with beta 4.
Elife, 8, 2019
|
|
6UQG
 
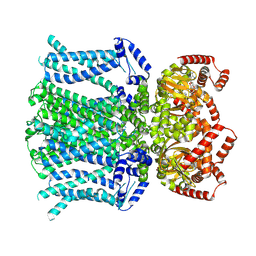 | | Human HCN1 channel Y289D mutant | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 | | Authors: | Lee, C.-H, MacKinnon, R. | | Deposit date: | 2019-10-19 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Voltage Sensor Movements during Hyperpolarization in the HCN Channel.
Cell, 179, 2019
|
|
6V35
 
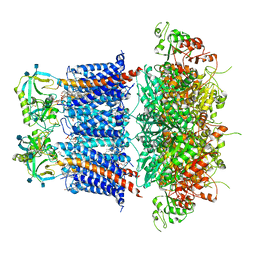 | | Cryo-EM structure of Ca2+-free hsSlo1-beta4 channel complex | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Tao, X, MacKinnon, R. | | Deposit date: | 2019-11-25 | | Release date: | 2019-12-25 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular structures of the human Slo1 K + channel in complex with beta 4.
Elife, 8, 2019
|
|
6V22
 
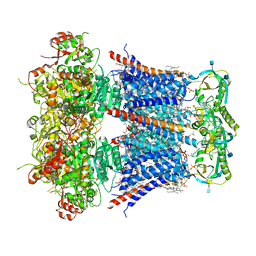 | | Cryo-EM structure of Ca2+-bound hsSlo1-beta4 channel complex | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Tao, X, MacKinnon, R. | | Deposit date: | 2019-11-22 | | Release date: | 2019-12-25 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular structures of the human Slo1 K + channel in complex with beta 4.
Elife, 8, 2019
|
|
5TQQ
 
 | |
6V38
 
 | | Cryo-EM structure of Ca2+-bound hsSlo1 channel | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CALCIUM ION, CHOLESTEROL, ... | | Authors: | Tao, X, MacKinnon, R. | | Deposit date: | 2019-11-25 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Molecular structures of the human Slo1 K + channel in complex with beta 4.
Elife, 8, 2019
|
|
5U6P
 
 | |
1J95
 
 | |
1JVM
 
 | |
1ID1
 
 | | CRYSTAL STRUCTURE OF THE RCK DOMAIN FROM E.COLI POTASSIUM CHANNEL | | Descriptor: | PUTATIVE POTASSIUM CHANNEL PROTEIN | | Authors: | Jiang, Y, Pico, A, Cadene, M, Chait, B.T, MacKinnon, R. | | Deposit date: | 2001-04-02 | | Release date: | 2001-04-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the RCK domain from the E. coli K+ channel and demonstration of its presence in the human BK channel.
Neuron, 29, 2001
|
|
1K4D
 
 | | Potassium Channel KcsA-Fab complex in low concentration of K+ | | Descriptor: | DIACYL GLYCEROL, NONAN-1-OL, POTASSIUM ION, ... | | Authors: | Zhou, Y, Morais-Cabral, J.H, Kaufman, A, MacKinnon, R. | | Deposit date: | 2001-10-07 | | Release date: | 2001-11-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 A resolution.
Nature, 414, 2001
|
|
1K4C
 
 | | Potassium Channel KcsA-Fab complex in high concentration of K+ | | Descriptor: | DIACYL GLYCEROL, NONAN-1-OL, POTASSIUM ION, ... | | Authors: | Zhou, Y, Morais-Cabral, J.H, Kaufman, A, MacKinnon, R. | | Deposit date: | 2001-10-07 | | Release date: | 2001-11-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 A resolution.
Nature, 414, 2001
|
|
3SPJ
 
 | |
3SYQ
 
 | | Crystal structure of the G protein-gated inward rectifier K+ channel GIRK2 (Kir3.2) R201A mutant in complex with PIP2 | | Descriptor: | G protein-activated inward rectifier potassium channel 2, POTASSIUM ION, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Whorton, M.R, MacKinnon, R. | | Deposit date: | 2011-07-18 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.44 Å) | | Cite: | Crystal Structure of the Mammalian GIRK2 K(+) Channel and Gating Regulation by G Proteins, PIP(2), and Sodium.
Cell(Cambridge,Mass.), 147, 2011
|
|
3U6N
 
 | | Open Structure of the BK channel Gating Ring | | Descriptor: | CALCIUM ION, High-Conductance Ca2+-Activated K+ Channel protein | | Authors: | Yuan, P, MacKinnon, R. | | Deposit date: | 2011-10-12 | | Release date: | 2011-12-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.61 Å) | | Cite: | Open structure of the Ca(2+) gating ring in the high-conductance Ca(2+)-activated K(+) channel.
Nature, 481, 2011
|
|
3SPG
 
 | | Inward rectifier potassium channel Kir2.2 R186A mutant in complex with PIP2 | | Descriptor: | Inward-rectifier K+ channel Kir2.2, POTASSIUM ION, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Hansen, S.B, Tao, X, MacKinnon, R. | | Deposit date: | 2011-07-01 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.613 Å) | | Cite: | Structural basis of PIP(2) activation of the classical inward rectifier K(+) channel Kir2.2.
Nature, 477, 2011
|
|
3SYP
 
 | |
3SYO
 
 | | Crystal structure of the G protein-gated inward rectifier K+ channel GIRK2 (Kir3.2) in complex with sodium | | Descriptor: | G protein-activated inward rectifier potassium channel 2, POTASSIUM ION, SODIUM ION | | Authors: | Whorton, M.R, MacKinnon, R. | | Deposit date: | 2011-07-18 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.54 Å) | | Cite: | Crystal Structure of the Mammalian GIRK2 K(+) Channel and Gating Regulation by G Proteins, PIP(2), and Sodium.
Cell(Cambridge,Mass.), 147, 2011
|
|
