7RX1
 
 | |
7S2Z
 
 | |
3ZLF
 
 | | Structure of group A Streptococcal enolase K312A mutant | | Descriptor: | ENOLASE, PHOSPHATE ION | | Authors: | Cork, A.J, Ericsson, D.J, Law, R.H.P, Casey, L.W, Valkov, E, Bertozzi, C, Stamp, A, Aquilina, J.A, Whisstock, J.C, Walker, M.J, Kobe, B. | | Deposit date: | 2013-01-31 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Stability of the Octameric Structure Affects Plasminogen-Binding Capacity of Streptococcal Enolase.
Plos One, 10, 2015
|
|
7MGQ
 
 | | AICAR transformylase/IMP cyclohydrolase (ATIC) is essential for de novo purine biosynthesis and infection by Cryptococcus neoformans | | Descriptor: | 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase, MAGNESIUM ION | | Authors: | Wizrah, M.S, Chua, S.M.H, Luo, Z, Manik, M.K, Pan, M, Whyte, J.M, Robertson, A.B, Kappler, U, Kobe, B, Fraser, J.A. | | Deposit date: | 2021-04-13 | | Release date: | 2022-04-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | AICAR transformylase/IMP cyclohydrolase (ATIC) is essential for de novo purine biosynthesis and infection by Cryptococcus neoformans.
J.Biol.Chem., 298, 2022
|
|
7NAK
 
 | | Cryo-EM structure of activated human SARM1 in complex with NMN and 1AD (TIR:1AD) | | Descriptor: | NAD(+) hydrolase SARM1, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-(5-iodanylisoquinolin-2-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Kerry, P.S, Nanson, J.D, Adams, S, Cunnea, K, Bosanac, T, Kobe, B, Hughes, R.O, Ve, T. | | Deposit date: | 2021-06-21 | | Release date: | 2022-03-23 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of SARM1 activation, substrate recognition, and inhibition by small molecules.
Mol.Cell, 82, 2022
|
|
7NAL
 
 | | Cryo-EM structure of activated human SARM1 in complex with NMN and 1AD (ARM and SAM domains) | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, NAD(+) hydrolase SARM1 | | Authors: | Kerry, P.S, Nanson, J.D, Adams, S, Cunnea, K, Bosanac, T, Kobe, B, Hughes, R.O, Ve, T. | | Deposit date: | 2021-06-21 | | Release date: | 2022-03-23 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of SARM1 activation, substrate recognition, and inhibition by small molecules.
Mol.Cell, 82, 2022
|
|
7MZQ
 
 | | Crystal structure of the UcaD lectin-binding domain in complex with fucose | | Descriptor: | CHLORIDE ION, Fimbrial adhesin UcaD, beta-L-fucopyranose | | Authors: | Ve, T, Lo, A.W, Schembri, M.A, Kobe, B. | | Deposit date: | 2021-05-24 | | Release date: | 2022-07-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Ucl fimbriae regulation and glycan receptor specificity contribute to gut colonisation by extra-intestinal pathogenic Escherichia coli.
Plos Pathog., 18, 2022
|
|
7MZO
 
 | | Crystal structure of the UcaD lectin-binding domain | | Descriptor: | CHLORIDE ION, Fimbrial adhesin UcaD | | Authors: | Ve, T, Lo, A.W, Schembri, M.A, Kobe, B. | | Deposit date: | 2021-05-24 | | Release date: | 2022-07-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Ucl fimbriae regulation and glycan receptor specificity contribute to gut colonisation by extra-intestinal pathogenic Escherichia coli.
Plos Pathog., 18, 2022
|
|
7MZP
 
 | | Crystal structure of the UclD lectin-binding domain | | Descriptor: | F17-like fimbril adhesin subunit UclD, IODIDE ION | | Authors: | Ve, T, Lo, A.W, Schembri, M.A, Kobe, B. | | Deposit date: | 2021-05-24 | | Release date: | 2022-07-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Ucl fimbriae regulation and glycan receptor specificity contribute to gut colonisation by extra-intestinal pathogenic Escherichia coli.
Plos Pathog., 18, 2022
|
|
7MZS
 
 | | Crystal structure of the UcaD lectin-binding domain in complex with galactose | | Descriptor: | CHLORIDE ION, Fimbrial adhesin UcaD, alpha-D-galactopyranose | | Authors: | Ve, T, Lo, A.W, Schembri, M.A, Kobe, B. | | Deposit date: | 2021-05-24 | | Release date: | 2022-07-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Ucl fimbriae regulation and glycan receptor specificity contribute to gut colonisation by extra-intestinal pathogenic Escherichia coli.
Plos Pathog., 18, 2022
|
|
7MZR
 
 | | Crystal structure of the UcaD lectin-binding domain in complex with glucose | | Descriptor: | CHLORIDE ION, Fimbrial adhesin UcaD, beta-D-glucopyranose | | Authors: | Ve, T, Lo, A.W, Schembri, M.A, Kobe, B. | | Deposit date: | 2021-05-24 | | Release date: | 2022-07-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Ucl fimbriae regulation and glycan receptor specificity contribute to gut colonisation by extra-intestinal pathogenic Escherichia coli.
Plos Pathog., 18, 2022
|
|
7L6W
 
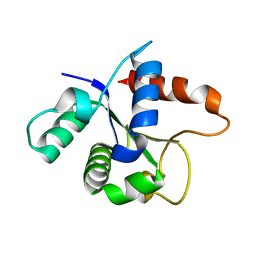 | | SFX structure of the MyD88 TIR domain higher-order assembly | | Descriptor: | Myeloid differentiation primary response protein MyD88 | | Authors: | Clabbers, M.T.B, Holmes, S, Muusse, T.W, Vajjhala, P, Thygesen, S.J, Malde, A.K, Hunter, D.J.B, Croll, T.I, Flueckiger, L, Nanson, J.D, Rahaman, M.H, Aquila, A, Hunter, M.S, Liang, M, Yoon, C.H, Zhao, J, Zatsepin, N.A, Abbey, B, Sierecki, E, Gambin, Y, Stacey, K.J, Darmanin, C, Kobe, B, Xu, H, Ve, T. | | Deposit date: | 2020-12-24 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | MyD88 TIR domain higher-order assembly interactions revealed by microcrystal electron diffraction and serial femtosecond crystallography.
Nat Commun, 12, 2021
|
|
4BOF
 
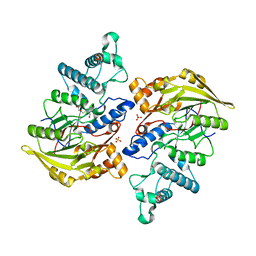 | | Crystal structure of arginine deiminase from group A streptococcus | | Descriptor: | ARGININE DEIMINASE, SULFATE ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Henningham, A, Ericsson, D.J, Langer, K, Casey, L, Jovcevski, B, Chhatwal, G.S, Aquilina, J.A, Batzloff, M.R, Kobe, B, Walker, M.J. | | Deposit date: | 2013-05-20 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structure-informed design of an enzymatically inactive vaccine component for group A Streptococcus.
MBio, 4, 2013
|
|
7L5S
 
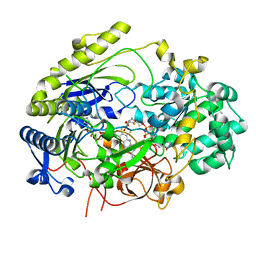 | | Crystal Structure of Haemophilus influenzae MtsZ at pH 5.5 | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, MOLYBDENUM ATOM, OXYGEN ATOM, ... | | Authors: | Struwe, M.A, Luo, Z, Kappler, U, Kobe, B. | | Deposit date: | 2020-12-22 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.089 Å) | | Cite: | Active site architecture reveals coordination sphere flexibility and specificity determinants in a group of closely related molybdoenzymes.
J.Biol.Chem., 296, 2021
|
|
7L5I
 
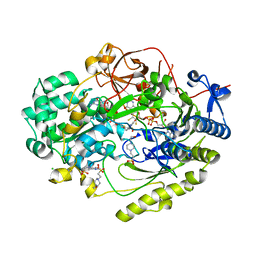 | | Crystal Structure of Haemophilus influenzae MtsZ at pH 7.0 | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Struwe, M.A, Luo, Z, Kappler, U, Kobe, B. | | Deposit date: | 2020-12-22 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.733 Å) | | Cite: | Active site architecture reveals coordination sphere flexibility and specificity determinants in a group of closely related molybdoenzymes.
J.Biol.Chem., 296, 2021
|
|
3SPS
 
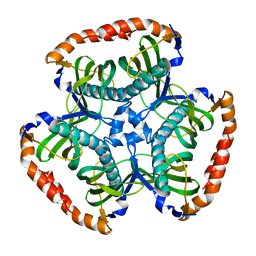 | |
3UL0
 
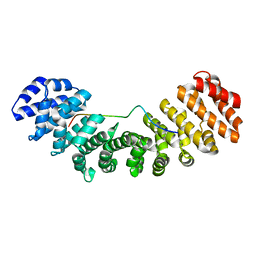 | | Mouse importin alpha: mouse CBP80Y8D cNLS complex | | Descriptor: | Importin subunit alpha-2, Nuclear cap-binding protein subunit 1 | | Authors: | Marfori, M, Forwood, J.K, Lonhienne, T.G, Kobe, B. | | Deposit date: | 2011-11-10 | | Release date: | 2012-10-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of High-Affinity Nuclear Localization Signal Interactions with Importin-alpha
Traffic, 13, 2012
|
|
3UX3
 
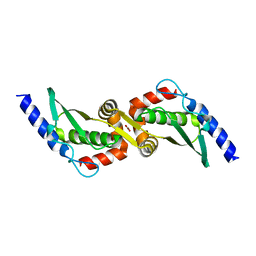 | | Crystal Structure of Domain-Swapped Fam96a minor dimer | | Descriptor: | ACETATE ION, MIP18 family protein FAM96A, ZINC ION | | Authors: | Chen, K.-E, Kobe, B, Martin, J.L. | | Deposit date: | 2011-12-03 | | Release date: | 2012-05-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The mammalian DUF59 protein Fam96a forms two distinct types of domain-swapped dimer.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3UX2
 
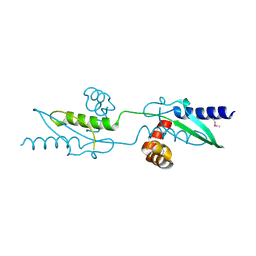 | |
3UKW
 
 | | Mouse importin alpha: Bimax1 peptide complex | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Bimax1 peptide, Importin subunit alpha-2 | | Authors: | Marfori, M, Forwood, J.K, Lonhienne, T.G, Kobe, B. | | Deposit date: | 2011-11-10 | | Release date: | 2012-10-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis of High-Affinity Nuclear Localization Signal Interactions with Importin-alpha
Traffic, 13, 2012
|
|
3UKX
 
 | | Mouse importin alpha: Bimax2 peptide complex | | Descriptor: | Bimax2 peptide, Importin subunit alpha-2 | | Authors: | Marfori, M, Forwood, J.K, Lonhienne, T.G, Kobe, B. | | Deposit date: | 2011-11-10 | | Release date: | 2012-10-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis of High-Affinity Nuclear Localization Signal Interactions with Importin-alpha
Traffic, 13, 2012
|
|
3UL1
 
 | | Mouse importin alpha: nucleoplasmin cNLS peptide complex | | Descriptor: | Importin subunit alpha-2, Nucleoplasmin | | Authors: | Marfori, M, Forwood, J.K, Lonhienne, T.G, Kobe, B. | | Deposit date: | 2011-11-10 | | Release date: | 2012-10-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of High-Affinity Nuclear Localization Signal Interactions with Importin-alpha
Traffic, 13, 2012
|
|
3UKZ
 
 | | Mouse importin alpha: mouse CBP80 cNLS complex | | Descriptor: | Importin subunit alpha-2, Nuclear cap-binding protein subunit 1 | | Authors: | Marfori, M, Forwood, J.K, Lonhienne, T.G, Kobe, B. | | Deposit date: | 2011-11-10 | | Release date: | 2012-10-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of High-Affinity Nuclear Localization Signal Interactions with Importin-alpha
Traffic, 13, 2012
|
|
3UKY
 
 | | Mouse importin alpha: yeast CBP80 cNLS complex | | Descriptor: | Importin subunit alpha-2, Nuclear cap-binding protein complex subunit 1 | | Authors: | Marfori, M, Forwood, J.K, Lonhienne, T.G, Kobe, B. | | Deposit date: | 2011-11-10 | | Release date: | 2012-10-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis of High-Affinity Nuclear Localization Signal Interactions with Importin-alpha
Traffic, 13, 2012
|
|
3ZIQ
 
 | | minor-site specific NLS (B6) | | Descriptor: | B6NLS, IMPORTIN SUBUNIT ALPHA-2 | | Authors: | Chang, C.-W, Counago, R.M, Williams, S.J, Kobe, B. | | Deposit date: | 2013-01-10 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Distinctive Conformation of Minor Site-Specific Nuclear Localization Signals Bound to Importin-Alpha
Traffic, 14, 2013
|
|
