8GSV
 
 | | Crystal structure of human BAK in complex with the Pxt1 BH3 domain | | Descriptor: | Bcl-2 homologous antagonist/killer, Peroxisomal testis-specific protein 1 | | Authors: | Lim, D, Ku, B. | | Deposit date: | 2022-09-07 | | Release date: | 2023-05-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for proapoptotic activation of Bak by the noncanonical BH3-only protein Pxt1.
Plos Biol., 21, 2023
|
|
1IZY
 
 | | Crystal structure of Hsp31 | | Descriptor: | Hsp31 | | Authors: | Cha, S.S, Lee, S.J. | | Deposit date: | 2002-10-16 | | Release date: | 2003-10-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of human DJ-1 and Escherichia coli Hsp31, which share an evolutionarily conserved domain
J.Biol.Chem., 278, 2003
|
|
1IZZ
 
 | | Crystal structure of Hsp31 | | Descriptor: | Hsp31 | | Authors: | Cha, S.S, Lee, S.J. | | Deposit date: | 2002-10-16 | | Release date: | 2003-10-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal structures of human DJ-1 and Escherichia coli Hsp31, which share an evolutionarily conserved domain
J.Biol.Chem., 278, 2003
|
|
4QXZ
 
 | |
5GOT
 
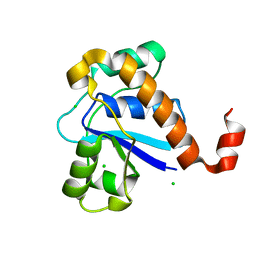 | | Crystal structure of SP-PTP, low molecular weight protein tyrosine phosphatase from Streptococcus pyogenes | | Descriptor: | CHLORIDE ION, Low molecular weight phosphotyrosine phosphatase family protein | | Authors: | Ku, B, Keum, C.W, KIim, S.J. | | Deposit date: | 2016-07-29 | | Release date: | 2016-09-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Crystal structure of SP-PTP, a low molecular weight protein tyrosine phosphatase from Streptococcus pyogenes
Biochem.Biophys.Res.Commun., 478, 2016
|
|
1J42
 
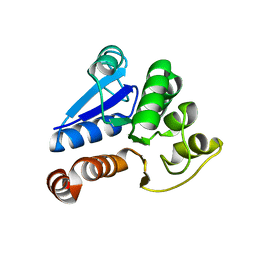 | | Crystal Structure of Human DJ-1 | | Descriptor: | RNA-binding protein regulatory subunit | | Authors: | Cha, S.S. | | Deposit date: | 2003-02-26 | | Release date: | 2004-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of human DJ-1 and Escherichia coli Hsp31, which share an evolutionarily conserved domain.
J.Biol.Chem., 278, 2003
|
|
5Y3D
 
 | |
5CSS
 
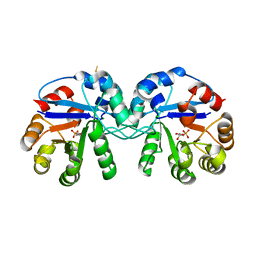 | | Crystal structure of triosephosphate isomerase from Thermoplasma acidophilum with glycerol 3-phosphate | | Descriptor: | CHLORIDE ION, SN-GLYCEROL-3-PHOSPHATE, Triosephosphate isomerase | | Authors: | Park, S.H, Kim, H.S, Song, M.K, Kim, K.R, Park, J.S, Han, B.W. | | Deposit date: | 2015-07-23 | | Release date: | 2016-06-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structure and Stability of the Dimeric Triosephosphate Isomerase from the Thermophilic Archaeon Thermoplasma acidophilum.
Plos One, 10, 2015
|
|
5C16
 
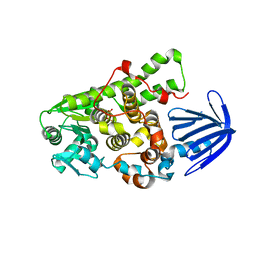 | | Myotubularin-related proetin 1 | | Descriptor: | Myotubularin-related protein 1, PHOSPHATE ION | | Authors: | Lee, B.I, Bong, S.M. | | Deposit date: | 2015-06-13 | | Release date: | 2016-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal Structure of Human Myotubularin-Related Protein 1 Provides Insight into the Structural Basis of Substrate Specificity
Plos One, 11, 2016
|
|
2EH7
 
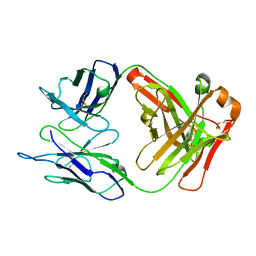 | | Crystal structure of humanized KR127 FAB | | Descriptor: | HUMANIZED KR127 FAB, HEAVY CHAIN, LIGHT CHAIN | | Authors: | Chi, S.-W, Kim, S.-J, Maeng, C.-Y, Hong, H.J, Ryu, S.-E. | | Deposit date: | 2007-03-05 | | Release date: | 2008-01-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Broadly neutralizing anti-hepatitis B virus antibody reveals a complementarity determining region H3 lid-opening mechanism
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3CMJ
 
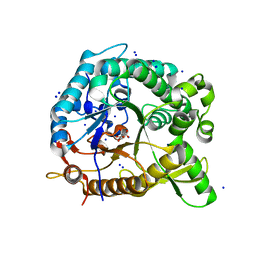 | |
2EH8
 
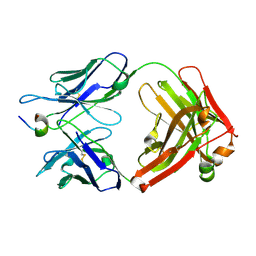 | | Crystal structure of the complex of humanized KR127 fab and PRES1 peptide epitope | | Descriptor: | HUMANIZED KR127 FAB, HEAVY CHAIN, LIGHT CHAIN, ... | | Authors: | Chi, S.-W, Kim, S.-J, Maeng, C.-Y, Hong, H.J, Ryu, S.-E. | | Deposit date: | 2007-03-05 | | Release date: | 2008-01-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Broadly neutralizing anti-hepatitis B virus antibody reveals a complementarity determining region H3 lid-opening mechanism
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
8IGC
 
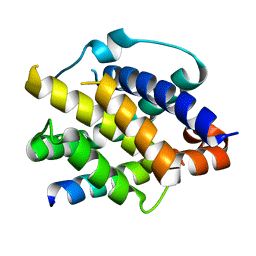 | | Crystal structure of Bak bound to Bnip5 BH3 | | Descriptor: | Bcl-2 homologous antagonist/killer, Protein BNIP5 | | Authors: | Ku, B, Lim, D. | | Deposit date: | 2023-02-20 | | Release date: | 2023-09-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.697 Å) | | Cite: | Crystal structure of Bak bound to the BH3 domain of Bnip5, a noncanonical BH3 domain-containing protein.
Proteins, 92, 2024
|
|
5CSR
 
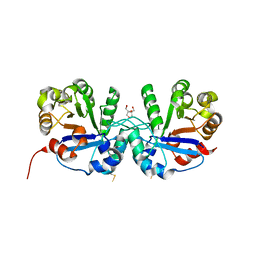 | | Crystal structure of triosephosphate isomerase from Thermoplasma acidophilium | | Descriptor: | CHLORIDE ION, GLYCEROL, Triosephosphate isomerase | | Authors: | Park, S.H, Kim, H.S, Song, M.K, Park, H.S, Han, B.W. | | Deposit date: | 2015-07-23 | | Release date: | 2016-06-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure and Stability of the Dimeric Triosephosphate Isomerase from the Thermophilic Archaeon Thermoplasma acidophilum.
Plos One, 10, 2015
|
|
8JOY
 
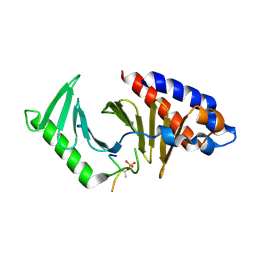 | | Plk1 polo-box domain bound to HPV4 L2 residues 251-257 with pThr255 | | Descriptor: | Peptide from Minor capsid protein L2, Serine/threonine-protein kinase PLK1 | | Authors: | Ku, B, Jung, S. | | Deposit date: | 2023-06-09 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal Structures of Plk1 Polo-Box Domain Bound to the Human Papillomavirus Minor Capsid Protein L2-Derived Peptide.
J.Microbiol, 61, 2023
|
|
8JOQ
 
 | |
2INV
 
 | |
2INU
 
 | |
4RCA
 
 | | Crystal structure of human PTPdelta and human Slitrk1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Receptor-type tyrosine-protein phosphatase delta, SLIT and NTRK-like protein 1, ... | | Authors: | Kim, H.M, Park, B.S, Kim, D, Lee, S.G. | | Deposit date: | 2014-09-15 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9908 Å) | | Cite: | Structural basis for LAR-RPTP/Slitrk complex-mediated synaptic adhesion.
Nat Commun, 5, 2014
|
|
4RCW
 
 | |
3K1J
 
 | | Crystal structure of Lon protease from Thermococcus onnurineus NA1 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Cha, S.S, An, Y.J. | | Deposit date: | 2009-09-28 | | Release date: | 2010-09-22 | | Last modified: | 2014-02-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Lon protease: molecular architecture of gated entry to a sequestered degradation chamber
Embo J., 29, 2010
|
|
5J1M
 
 | | Crystal structure of Csd1-Csd2 dimer II | | Descriptor: | ToxR-activated gene (TagE), ZINC ION | | Authors: | An, D.R, Suh, S.W. | | Deposit date: | 2016-03-29 | | Release date: | 2016-10-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis of the Heterodimer Formation between Cell Shape-Determining Proteins Csd1 and Csd2 from Helicobacter pylori
Plos One, 11, 2016
|
|
5J1K
 
 | | Crystal structure of Csd2-Csd2 dimer | | Descriptor: | GLYCEROL, ToxR-activated gene (TagE) | | Authors: | An, D.R, Suh, S.W. | | Deposit date: | 2016-03-29 | | Release date: | 2016-10-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural Basis of the Heterodimer Formation between Cell Shape-Determining Proteins Csd1 and Csd2 from Helicobacter pylori
Plos One, 11, 2016
|
|
5J1L
 
 | | Crystal structure of Csd1-Csd2 dimer I | | Descriptor: | ToxR-activated gene (TagE), ZINC ION | | Authors: | An, D.R, Suh, S.W. | | Deposit date: | 2016-03-29 | | Release date: | 2016-10-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural Basis of the Heterodimer Formation between Cell Shape-Determining Proteins Csd1 and Csd2 from Helicobacter pylori
Plos One, 11, 2016
|
|
1ZKJ
 
 | | Structural Basis for the Extended Substrate Spectrum of CMY-10, a Plasmid-Encoded Class C beta-lactamase | | Descriptor: | ACETIC ACID, ZINC ION, extended-spectrum beta-lactamase | | Authors: | Cha, S.S, Jung, H.I, An, Y.J, Lee, S.H. | | Deposit date: | 2005-05-03 | | Release date: | 2006-04-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for the extended substrate spectrum of CMY-10, a plasmid-encoded class C beta-lactamase.
Mol.Microbiol., 60, 2006
|
|
