1BT3
 
 | | CATECHOL OXIDASE FROM IPOMOEA BATATAS (SWEET POTATOES) IN THE NATIVE CU(II)-CU(II) STATE | | Descriptor: | CU-O-CU LINKAGE, PROTEIN (CATECHOL OXIDASE) | | Authors: | Klabunde, T, Eicken, C, Sacchettini, J.C, Krebs, B. | | Deposit date: | 1998-09-02 | | Release date: | 1999-09-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a plant catechol oxidase containing a dicopper center.
Nat.Struct.Biol., 5, 1998
|
|
1BUG
 
 | | CATECHOL OXIDASE FROM IPOMOEA BATATAS (SWEET POTATOES)-INHIBITOR COMPLEX WITH PHENYLTHIOUREA (PTU) | | Descriptor: | COPPER (II) ION, N-PHENYLTHIOUREA, PROTEIN (CATECHOL OXIDASE) | | Authors: | Klabunde, T, Eicken, C, Sacchettini, J.C, Krebs, B. | | Deposit date: | 1998-09-03 | | Release date: | 1999-09-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a plant catechol oxidase containing a dicopper center.
Nat.Struct.Biol., 5, 1998
|
|
1COZ
 
 | | CTP:GLYCEROL-3-PHOSPHATE CYTIDYLYLTRANSFERASE FROM BACILLUS SUBTILIS | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, PROTEIN (GLYCEROL-3-PHOSPHATE CYTIDYLYLTRANSFERASE) | | Authors: | Weber, C.H, Park, Y.S, Sanker, S, Kent, C, Ludwig, M.L. | | Deposit date: | 1999-05-29 | | Release date: | 1999-10-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A prototypical cytidylyltransferase: CTP:glycerol-3-phosphate cytidylyltransferase from bacillus subtilis.
Structure Fold.Des., 7, 1999
|
|
1BT1
 
 | | CATECHOL OXIDASE FROM IPOMOEA BATATAS (SWEET POTATOES) IN THE NATIVE CU(II)-CU(II) STATE | | Descriptor: | CU-O-CU LINKAGE, PROTEIN (CATECHOL OXIDASE) | | Authors: | Klabunde, T, Eicken, C, Sacchettini, J.C, Krebs, B. | | Deposit date: | 1998-09-02 | | Release date: | 1999-09-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a plant catechol oxidase containing a dicopper center.
Nat.Struct.Biol., 5, 1998
|
|
1BT2
 
 | | CATECHOL OXIDASE FROM IPOMOEA BATATAS (SWEET POTATOES) IN THE REDUCED CU(I)-CU(I) STATE | | Descriptor: | CU-O-CU LINKAGE, PROTEIN (CATECHOL OXIDASE) | | Authors: | Klabunde, T, Eicken, C, Sacchettini, J.C, Krebs, B. | | Deposit date: | 1998-09-02 | | Release date: | 1999-09-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a plant catechol oxidase containing a dicopper center.
Nat.Struct.Biol., 5, 1998
|
|
1Z41
 
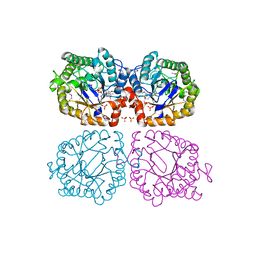 | | Crystal structure of oxidized YqjM from Bacillus subtilis | | Descriptor: | FLAVIN MONONUCLEOTIDE, Probable NADH-dependent flavin oxidoreductase yqjM, SULFATE ION | | Authors: | Kitzing, K, Fitzpatrick, T.B, Wilken, C, Sawa, J, Bourenkov, G.P, Macheroux, P, Clausen, T. | | Deposit date: | 2005-03-15 | | Release date: | 2005-05-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The 1.3 A Crystal Structure of the Flavoprotein YqjM Reveals a Novel Class of Old Yellow Enzymes
J.Biol.Chem., 280, 2005
|
|
1Z48
 
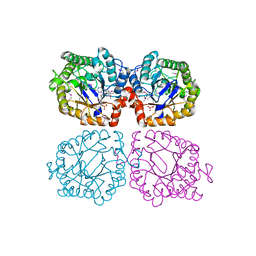 | | Crystal structure of reduced YqjM from Bacillus subtilis | | Descriptor: | FLAVIN MONONUCLEOTIDE, Probable NADH-dependent flavin oxidoreductase yqjM | | Authors: | Kitzing, K, Fitzpatrick, T.B, Wilken, C, Sawa, J, Bourenkov, G.P, Macheroux, P, Clausen, T. | | Deposit date: | 2005-03-15 | | Release date: | 2005-05-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The 1.3 A Crystal Structure of the Flavoprotein YqjM Reveals a Novel Class of Old Yellow Enzymes
J.Biol.Chem., 280, 2005
|
|
1Z42
 
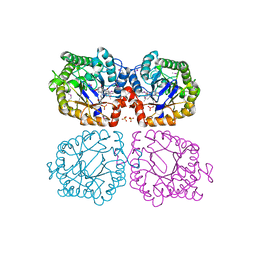 | | Crystal structure of oxidized YqjM from Bacillus subtilis complexed with p-hydroxybenzaldehyde | | Descriptor: | FLAVIN MONONUCLEOTIDE, P-HYDROXYBENZALDEHYDE, Probable NADH-dependent flavin oxidoreductase yqjM, ... | | Authors: | Kitzing, K, Fitzpatrick, T.B, Wilken, C, Sawa, J, Bourenkov, G.P, Macheroux, P, Clausen, T. | | Deposit date: | 2005-03-15 | | Release date: | 2005-05-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The 1.3 A Crystal Structure of the Flavoprotein YqjM Reveals a Novel Class of Old Yellow Enzymes
J.Biol.Chem., 280, 2005
|
|
2LBH
 
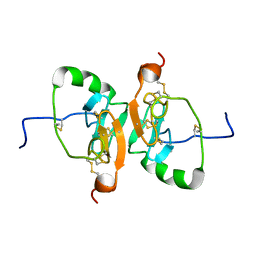 | | Solution Structure of the Dimeric Form of a Unliganded Bovine Neurophysin, Minimized Average Structure | | Descriptor: | Neurophysin 1 | | Authors: | Lee, H, Naik, M, Nguyen, T, Bracken, C, Breslow, E. | | Deposit date: | 2011-03-31 | | Release date: | 2012-04-04 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of the Dimerization-Induced Increase in Neurophysin-Hormone Affinity: Interplay of Inter-Domain and Inter-Subunit Interactions
To be Published
|
|
2LBN
 
 | |
2NMM
 
 | | Crystal structure of human phosphohistidine phosphatase. Northeast Structural Genomics Consortium target HR1409 | | Descriptor: | 14 kDa phosphohistidine phosphatase, SULFATE ION | | Authors: | Kuzin, A.P, Abashidze, M, Forouhar, F, Seetharaman, J, Kent, C, Fang, Y, Cunningham, K, Conover, K, Ma, L.C, Xiao, R, Acton, T, Montelione, G, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-10-22 | | Release date: | 2007-01-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of human phosphohistidine phosphatase. Northeast Structural Genomics Consortium target HR1409
To be Published
|
|
5NPT
 
 | | Structure of the N-terminal domain of the yeast telomerase reverse transcriptase | | Descriptor: | Telomerase reverse transcriptase | | Authors: | Rodina, E.V, Lebedev, A.A, Hakanpaa, J, Hackenberg, C, Petrova, O.A, Zvereva, M.I, Dontsova, O.A, Lamzin, V.S. | | Deposit date: | 2017-04-18 | | Release date: | 2017-12-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and function of the N-terminal domain of the yeast telomerase reverse transcriptase.
Nucleic Acids Res., 46, 2018
|
|
6HON
 
 | | Drosophila NOT4 CBM peptide bound to human CAF40 | | Descriptor: | CCR4-NOT transcription complex subunit 4, isoform L, CCR4-NOT transcription complex subunit 9, ... | | Authors: | Raisch, T, Izaurralde, E, Weichenrieder, O. | | Deposit date: | 2018-09-17 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A conserved CAF40-binding motif in metazoan NOT4 mediates association with the CCR4-NOT complex.
Genes Dev., 33, 2019
|
|
6HOM
 
 | | Drosophila NOT4 CBM peptide bound to human CAF40 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CCR4-NOT transcription complex subunit 4, isoform L, ... | | Authors: | Raisch, T, Izaurralde, E, Weichenrieder, O. | | Deposit date: | 2018-09-17 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A conserved CAF40-binding motif in metazoan NOT4 mediates association with the CCR4-NOT complex.
Genes Dev., 33, 2019
|
|
6CHZ
 
 | | Estrogen Receptor Alpha Y537S bound to antagonist H3B-9224. | | Descriptor: | 1,2-ETHANEDIOL, 4-[(2-{4-[(1E)-1-(1H-indazol-5-yl)-2-phenylbut-1-en-1-yl]phenoxy}ethyl)amino]-N,N-dimethylbutanamide, Estrogen receptor | | Authors: | Larsen, N.A. | | Deposit date: | 2018-02-23 | | Release date: | 2018-03-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Discovery of Selective Estrogen Receptor Covalent Antagonists for the Treatment of ER alphaWTand ER alphaMUTBreast Cancer.
Cancer Discov, 8, 2018
|
|
5VB6
 
 | |
7ASL
 
 | |
7ASH
 
 | |
4RX0
 
 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM265 | | Descriptor: | 2-(1,1-difluoroethyl)-5-methyl-N-[4-(pentafluoro-lambda~6~-sulfanyl)phenyl][1,2,4]triazolo[1,5-a]pyrimidin-7-amine, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Deng, X, Phillips, M, Tomchick, D. | | Deposit date: | 2014-12-08 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A long-duration dihydroorotate dehydrogenase inhibitor (DSM265) for prevention and treatment of malaria.
Sci Transl Med, 7, 2015
|
|
5VND
 
 | | Crystal structure of FGFR1-Y563C (FGFR4 surrogate) covalently bound to H3B-6527 | | Descriptor: | 1,2-ETHANEDIOL, Fibroblast growth factor receptor 1, N-{2-[(6-{[(2,6-dichloro-3,5-dimethoxyphenyl)carbamoyl](methyl)amino}pyrimidin-4-yl)amino]-5-(4-ethylpiperazin-1-yl)phenyl}propanamide, ... | | Authors: | Tsai, J.H.C, Reynolds, D, Fekkes, P, Smith, P, Larsen, N.A. | | Deposit date: | 2017-04-30 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | H3B-6527 Is a Potent and Selective Inhibitor of FGFR4 in FGF19-Driven Hepatocellular Carcinoma.
Cancer Res., 77, 2017
|
|
4LWD
 
 | | Human CARMA1 CARD domain | | Descriptor: | Caspase recruitment domain-containing protein 11, MAGNESIUM ION, SULFATE ION | | Authors: | Zheng, C, Wu, H. | | Deposit date: | 2013-07-26 | | Release date: | 2013-10-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Structural Architecture of the CARMA1/Bcl10/MALT1 Signalosome: Nucleation-Induced Filamentous Assembly.
Mol.Cell, 51, 2013
|
|
6T28
 
 | | Crystal structure of human calmodulin-dependent protein kinase 1D (CAMK1D) bound to compound 19 (CS640) | | Descriptor: | 1,2-ETHANEDIOL, 2-[(3~{S})-3-azanylpiperidin-1-yl]-4-[[2,6-di(propan-2-yl)pyridin-4-yl]amino]pyrimidine-5-carboxamide, Calcium/calmodulin-dependent protein kinase type 1D, ... | | Authors: | Kraemer, A, Sorrell, F, Butterworth, S, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-08 | | Release date: | 2019-11-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of Highly Selective Inhibitors of Calmodulin-Dependent Kinases That Restore Insulin Sensitivity in the Diet-Induced Obesityin VivoMouse Model.
J.Med.Chem., 63, 2020
|
|
7M7V
 
 | | Crystal Structure of Mtb Pks13 Thioesterase domain in complex with Compound 6 | | Descriptor: | 5-hydroxy-2-(4-hydroxyphenyl)-N-methyl-4-[(2-oxa-6-azaspiro[3.4]octan-6-yl)methyl]-1-benzofuran-3-carboxamide, Polyketide synthase Pks13 | | Authors: | Aggarwal, A, Sacchettini, J.C. | | Deposit date: | 2021-03-29 | | Release date: | 2022-02-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Optimization of TAM16, a Benzofuran That Inhibits the Thioesterase Activity of Pks13; Evaluation toward a Preclinical Candidate for a Novel Antituberculosis Clinical Target.
J.Med.Chem., 65, 2022
|
|
6T29
 
 | | Crystal structure of human calmodulin-dependent protein kinase 1D (CAMK1D) bound to compound 18 (CS587) | | Descriptor: | 1,2-ETHANEDIOL, 2-[(3~{S})-3-azanylpiperidin-1-yl]-4-[[3,5-bis(2-cyanopropan-2-yl)phenyl]amino]pyrimidine-5-carboxamide, Calcium/calmodulin-dependent protein kinase type 1D, ... | | Authors: | Kraemer, A, Sorrell, F, Butterworth, S, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-08 | | Release date: | 2019-11-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.484 Å) | | Cite: | Discovery of Highly Selective Inhibitors of Calmodulin-Dependent Kinases That Restore Insulin Sensitivity in the Diet-Induced Obesityin VivoMouse Model.
J.Med.Chem., 63, 2020
|
|
3U67
 
 | | Crystal structure of the N-terminal domain of Hsp90 from Leishmania major(LmjF33.0312)in complex with ADP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, Heat shock protein 83-1, ... | | Authors: | Pizarro, J.C, Wernimont, A.K, Hutchinson, A, Mackenzie, F, Fairlamb, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Ferguson, M.A.J, Hui, R, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-10-12 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Exploring the Trypanosoma brucei Hsp83 potential as a target for structure guided drug design.
PLoS Negl Trop Dis, 7, 2013
|
|
