3TI3
 
 | | Crystal structure of 2009 pandemic H1N1 neuraminidase complexed with laninamivir | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-acetamido-2,6-anhydro-4-carbamimidamido-3,4,5-trideoxy-7-O-methyl-D-glycero-D-galacto-non-2-enonic acid, ... | | Authors: | Vavricka, C.J, Li, Q, Wu, Y, Qi, J, Wang, M, Liu, Y, Gao, F, Liu, J, Feng, E, He, J, Wang, J, Liu, H, Jiang, H, Gao, G.F. | | Deposit date: | 2011-08-20 | | Release date: | 2011-11-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and functional analysis of laninamivir and its octanoate prodrug reveals group specific mechanisms for influenza NA inhibition
Plos Pathog., 7, 2011
|
|
5BWO
 
 | | Crystal Structure of Human SIRT3 in Complex with a Palmitoyl H3K9 Peptide | | Descriptor: | NAD-dependent protein deacetylase sirtuin-3, mitochondrial, PALMITIC ACID, ... | | Authors: | Gai, W, Jiang, H, Liu, D. | | Deposit date: | 2015-06-08 | | Release date: | 2016-07-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.376 Å) | | Cite: | Crystal structures of SIRT3 reveal that the alpha 2-alpha 3 loop and alpha 3-helix affect the interaction with long-chain acyl lysine.
Febs Lett., 590, 2016
|
|
3TI5
 
 | | Crystal structure of 2009 pandemic H1N1 neuraminidase complexed with Zanamivir | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CALCIUM ION, ... | | Authors: | Vavricka, C.J, Li, Q, Wu, Y, Qi, J, Wang, M, Liu, Y, Gao, F, Liu, J, Feng, E, He, J, Wang, J, Liu, H, Jiang, H, Gao, G.F. | | Deposit date: | 2011-08-20 | | Release date: | 2011-11-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional analysis of laninamivir and its octanoate prodrug reveals group specific mechanisms for influenza NA inhibition
Plos Pathog., 7, 2011
|
|
3TI6
 
 | | Crystal structure of 2009 pandemic H1N1 neuraminidase complexed with oseltamivir | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Vavricka, C.J, Li, Q, Wu, Y, Qi, J, Wang, M, Liu, Y, Gao, F, Liu, J, Feng, E, He, J, Wang, J, Liu, H, Jiang, H, Gao, G.F. | | Deposit date: | 2011-08-20 | | Release date: | 2011-11-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural and functional analysis of laninamivir and its octanoate prodrug reveals group specific mechanisms for influenza NA inhibition
Plos Pathog., 7, 2011
|
|
3TI8
 
 | | Crystal structure of influenza A virus neuraminidase N5 complexed with laninamivir | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-acetamido-2,6-anhydro-4-carbamimidamido-3,4,5-trideoxy-7-O-methyl-D-glycero-D-galacto-non-2-enonic acid, ... | | Authors: | Vavricka, C.J, Li, Q, Wu, Y, Qi, J, Wang, M, Liu, Y, Gao, F, Liu, J, Feng, E, He, J, Wang, J, Liu, H, Jiang, H, Gao, G.F. | | Deposit date: | 2011-08-20 | | Release date: | 2011-11-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Structural and functional analysis of laninamivir and its octanoate prodrug reveals group specific mechanisms for influenza NA inhibition
Plos Pathog., 7, 2011
|
|
3HHS
 
 | | Crystal Structure of Manduca sexta prophenoloxidase | | Descriptor: | COPPER (II) ION, Phenoloxidase subunit 1, Phenoloxidase subunit 2 | | Authors: | Li, Y, Wang, Y, Jiang, H, Deng, J. | | Deposit date: | 2009-05-17 | | Release date: | 2009-09-29 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of Manduca sexta prophenoloxidase provides insights into the mechanism of type 3 copper enzymes.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2PVP
 
 | |
3FXO
 
 | | Crystal Structure of Human Protein phosphatase 1A (PPM1A) Bound with Phosphate at 1 mM of Mn2+ | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, Protein phosphatase 1A | | Authors: | Hu, T, Wang, L, Wang, K, Jiang, H, Shen, X. | | Deposit date: | 2009-01-21 | | Release date: | 2010-01-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the Mn2+-dependent activation of human PPM1A
To be published
|
|
3FXM
 
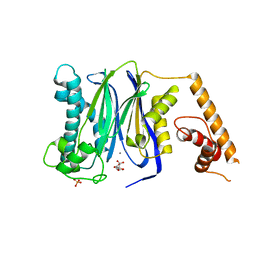 | | Crystal Structure of Human Protein phosphatase 1A (PPM1A) Bound with Citrate at 10 mM of Mn2+ | | Descriptor: | CITRATE ANION, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Hu, T, Wang, L, Wang, K, Jiang, H, Shen, X. | | Deposit date: | 2009-01-21 | | Release date: | 2010-01-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the Mn2+-dependent activation of human PPM1A
To be published
|
|
2PWX
 
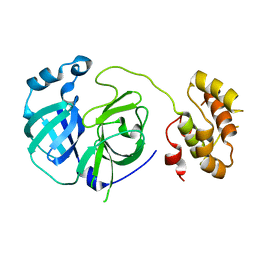 | | Crystal structure of G11A mutant of SARS-CoV 3C-like protease | | Descriptor: | 3C-like proteinase | | Authors: | Chen, S, Hu, T, Jiang, H, Shen, X. | | Deposit date: | 2007-05-14 | | Release date: | 2007-10-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mutation of Gly-11 on the dimer interface results in the complete crystallographic dimer dissociation of severe acute respiratory syndrome coronavirus 3C-like protease: crystal structure with molecular dynamics simulations.
J.Biol.Chem., 283, 2008
|
|
2RJG
 
 | | Crystal structure of biosynthetic alaine racemase from Escherichia coli | | Descriptor: | Alanine racemase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Wu, D, Hu, T, Zhang, L, Jiang, H, Shen, X. | | Deposit date: | 2007-10-15 | | Release date: | 2008-07-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Residues Asp164 and Glu165 at the substrate entryway function potently in substrate orientation of alanine racemase from E. coli: Enzymatic characterization with crystal structure analysis
Protein Sci., 17, 2008
|
|
2QGH
 
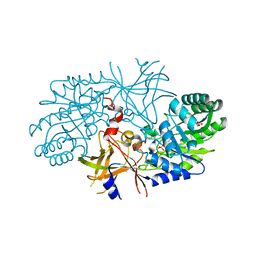 | | Crystal structure of diaminopimelate decarboxylase from Helicobacter pylori complexed with L-lysine | | Descriptor: | Diaminopimelate decarboxylase, GLYCEROL, LYSINE, ... | | Authors: | Hu, T, Wu, D, Jiang, H, Shen, X. | | Deposit date: | 2007-06-28 | | Release date: | 2008-05-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of diaminopimelate decarboxylase from Helicobacter pylori complexed with L-lysine
TO BE PUBLISHED
|
|
2RJH
 
 | | Crystal structure of biosynthetic alaine racemase in D-cycloserine-bound form from Escherichia coli | | Descriptor: | Alanine racemase, D-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-N,O-CYCLOSERYLAMIDE, SULFATE ION | | Authors: | Wu, D, Hu, T, Zhang, L, Jiang, H, Shen, X. | | Deposit date: | 2007-10-15 | | Release date: | 2008-07-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Residues Asp164 and Glu165 at the substrate entryway function potently in substrate orientation of alanine racemase from E. coli: Enzymatic characterization with crystal structure analysis
Protein Sci., 17, 2008
|
|
6LN2
 
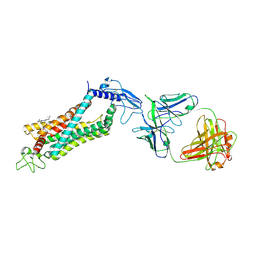 | | Crystal structure of full length human GLP1 receptor in complex with Fab fragment (Fab7F38) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab7F38_heavy chain, Fab7F38_light chain, ... | | Authors: | Wu, F, Yang, L, Hang, K, Laursen, M, Wu, L, Han, G.W, Ren, Q, Roed, N.K, Lin, G, Hanson, M, Jiang, H, Wang, M, Reedtz-Runge, S, Song, G, Stevens, R.C. | | Deposit date: | 2019-12-28 | | Release date: | 2020-03-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Full-length human GLP-1 receptor structure without orthosteric ligands.
Nat Commun, 11, 2020
|
|
2GLP
 
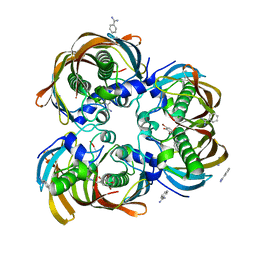 | | Crystal structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase(FabZ) from Helicobacter pylori complexed with compound 1 | | Descriptor: | (3R)-hydroxymyristoyl-acyl carrier protein dehydratase, BENZAMIDINE, CHLORIDE ION, ... | | Authors: | Zhang, L, Liu, W, Shen, X, Jiang, H. | | Deposit date: | 2006-04-05 | | Release date: | 2007-03-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural basis for catalytic and inhibitory mechanisms of beta-hydroxyacyl-acyl carrier protein dehydratase (FabZ).
J.Biol.Chem., 283, 2008
|
|
2GLV
 
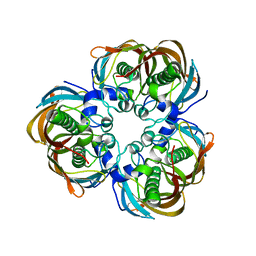 | | Crystal structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase(FabZ) mutant(Y100A) from Helicobacter pylori | | Descriptor: | (3R)-hydroxymyristoyl-acyl carrier protein dehydratase, CHLORIDE ION | | Authors: | Zhang, L, Liu, W, Shen, X, Jiang, H. | | Deposit date: | 2006-04-05 | | Release date: | 2007-03-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for catalytic and inhibitory mechanisms of beta-hydroxyacyl-acyl carrier protein dehydratase (FabZ).
J.Biol.Chem., 283, 2008
|
|
2GLL
 
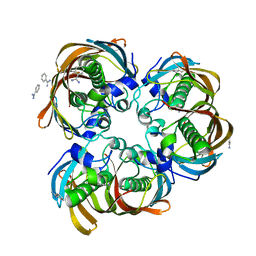 | | Crystal structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase(FabZ) from Helicobacter pylori | | Descriptor: | (3R)-hydroxymyristoyl-acyl carrier protein dehydratase, BENZAMIDINE, CHLORIDE ION | | Authors: | Zhang, L, Liu, W, Shen, X, Jiang, H. | | Deposit date: | 2006-04-05 | | Release date: | 2007-03-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for catalytic and inhibitory mechanisms of beta-hydroxyacyl-acyl carrier protein dehydratase (FabZ).
J.Biol.Chem., 283, 2008
|
|
2H1Y
 
 | |
2GLM
 
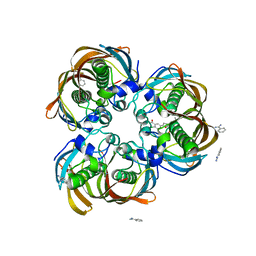 | | Crystal structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase(FabZ) from Helicobacter pylori complexed with Compound 2 | | Descriptor: | (3R)-hydroxymyristoyl-acyl carrier protein dehydratase, 2-CHLORO-5-(5-{(E)-[(2Z)-3-(2-METHOXYETHYL)-4-OXO-2-(PHENYLIMINO)-1,3-THIAZOLIDIN-5-YLIDENE]METHYL}-2-FURYL)BENZOIC ACID, BENZAMIDINE, ... | | Authors: | Zhang, L, Liu, W, Shen, X, Jiang, H. | | Deposit date: | 2006-04-05 | | Release date: | 2007-03-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for catalytic and inhibitory mechanisms of beta-hydroxyacyl-acyl carrier protein dehydratase (FabZ).
J.Biol.Chem., 283, 2008
|
|
4IAR
 
 | | Crystal structure of the chimeric protein of 5-HT1B-BRIL in complex with ergotamine (PSI Community Target) | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Chimera protein of human 5-hydroxytryptamine receptor 1B and E. Coli soluble cytochrome b562, Ergotamine | | Authors: | Wang, C, Jiang, Y, Ma, J, Wu, H, Wacker, D, Katritch, V, Han, G.W, Liu, W, Huang, X, Vardy, E, McCorvy, J.D, Gao, X, Zhou, E.X, Melcher, K, Zhang, C, Bai, F, Yang, H, Yang, L, Jiang, H, Roth, B.L, Cherezov, V, Stevens, R.C, Xu, H.E, GPCR Network (GPCR) | | Deposit date: | 2012-12-07 | | Release date: | 2013-03-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for molecular recognition at serotonin receptors.
Science, 340, 2013
|
|
3ST9
 
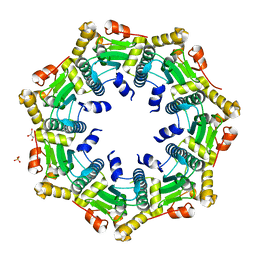 | | Crystal structure of ClpP in heptameric form from Staphylococcus aureus | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, CALCIUM ION, GLYCEROL, ... | | Authors: | Zhang, J, Ye, F, Lan, L, Jiang, H, Luo, C, Yang, C.-G. | | Deposit date: | 2011-07-09 | | Release date: | 2011-09-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural switching of Staphylococcus aureus Clp protease: a key to understanding protease dynamics
J.Biol.Chem., 286, 2011
|
|
3STA
 
 | | Crystal structure of ClpP in tetradecameric form from Staphylococcus aureus | | Descriptor: | ATP-dependent Clp protease proteolytic subunit | | Authors: | Zhang, J, Ye, F, Lan, L, Jiang, H, Luo, C, Yang, C.-G. | | Deposit date: | 2011-07-09 | | Release date: | 2011-09-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural switching of Staphylococcus aureus Clp protease: a key to understanding protease dynamics
J.Biol.Chem., 286, 2011
|
|
4ZJB
 
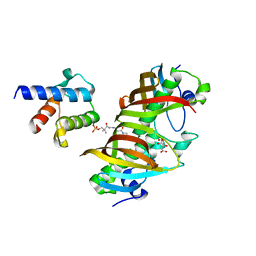 | | Crystal structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase(FabZ) in complex with holo-ACP from Helicobacter pylori | | Descriptor: | 3-hydroxyacyl-[acyl-carrier-protein] dehydratase FabZ, 4'-PHOSPHOPANTETHEINE, Acyl carrier protein, ... | | Authors: | Zhang, L, Zhang, L, Shen, X, Jiang, H. | | Deposit date: | 2015-04-29 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of FabZ-ACP complex reveals a dynamic seesaw-like catalytic mechanism of dehydratase in fatty acid biosynthesis.
Cell Res., 26, 2016
|
|
4IAQ
 
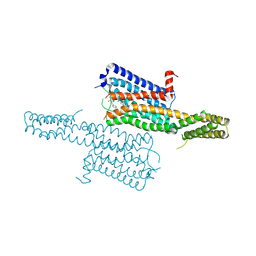 | | Crystal structure of the chimeric protein of 5-HT1B-BRIL in complex with dihydroergotamine (PSI Community Target) | | Descriptor: | Chimera protein of human 5-hydroxytryptamine receptor 1B and E. Coli soluble cytochrome b562, Dihydroergotamine | | Authors: | Wang, C, Jiang, Y, Ma, J, Wu, H, Wacker, D, Katritch, V, Han, G.W, Liu, W, Huang, X, Vardy, E, McCorvy, J.D, Gao, X, Zhou, E.X, Melcher, K, Zhang, C, Bai, F, Yang, H, Yang, L, Jiang, H, Roth, B.L, Cherezov, V, Stevens, R.C, Xu, H.E, GPCR Network (GPCR) | | Deposit date: | 2012-12-07 | | Release date: | 2013-03-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for molecular recognition at serotonin receptors.
Science, 340, 2013
|
|
5BWL
 
 | |
