5Y0N
 
 | |
5Y0Q
 
 | | Crystal structure of BsTmcAL bound with AMPCPP | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, UPF0348 protein B4417_3650 | | Authors: | Yamashita, S, Tomita, K. | | Deposit date: | 2017-07-18 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Acetate-dependent tRNA acetylation required for decoding fidelity in protein synthesis.
Nat. Chem. Biol., 14, 2018
|
|
5Y0O
 
 | | Crystal structure of apo BsTmcAL | | Descriptor: | UPF0348 protein B4417_3650 | | Authors: | Yamashita, S, Tomita, K. | | Deposit date: | 2017-07-18 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Acetate-dependent tRNA acetylation required for decoding fidelity in protein synthesis.
Nat. Chem. Biol., 14, 2018
|
|
5Y0T
 
 | |
5Y0S
 
 | |
5Y0R
 
 | |
8IE2
 
 | | Crystal structure of Lactiplantibacillus plantarum GlyRS | | Descriptor: | Glycine--tRNA ligase alpha subunit, Glycine--tRNA ligase beta subunit | | Authors: | Yamashita, S, Tomita, K. | | Deposit date: | 2023-02-15 | | Release date: | 2023-06-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Mechanism of tRNA recognition by heterotetrameric glycyl-tRNA synthetase from lactic acid bacteria.
J.Biochem., 174, 2023
|
|
7VNX
 
 | | Crystal structure of TkArkI | | Descriptor: | GUANOSINE, TkArkI | | Authors: | Yamashita, S, Minowa, K, Ohira, T, Suzuki, T, Tomita, K. | | Deposit date: | 2021-10-12 | | Release date: | 2022-05-04 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Reversible RNA phosphorylation stabilizes tRNA for cellular thermotolerance.
Nature, 605, 2022
|
|
7VNV
 
 | |
7VNW
 
 | |
7E5O
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with antibody NT-193 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NT-193 Heavy chain, NT-193 Light chain, ... | | Authors: | Kita, S, Onodera, T, Adachi, Y, Moriayma, S, Nomura, T, Tadokoro, T, Anraku, Y, Yumoto, K, Tian, C, Fukuhara, H, Suzuki, T, Tonouchi, K, Sasaki, J, Sun, L, Hashiguchi, T, Takahashi, Y, Maenaka, K. | | Deposit date: | 2021-02-19 | | Release date: | 2021-09-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A SARS-CoV-2 antibody broadly neutralizes SARS-related coronaviruses and variants by coordinated recognition of a virus-vulnerable site.
Immunity, 54, 2021
|
|
1SWG
 
 | | CIRCULAR PERMUTED STREPTAVIDIN E51/A46 IN COMPLEX WITH BIOTIN | | Descriptor: | BIOTIN, CIRCULARLY PERMUTED CORE-STREPTAVIDIN E51/A46 | | Authors: | Freitag, S, Chu, V, Le Trong, I, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1997-07-12 | | Release date: | 1998-07-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Thermodynamic and structural consequences of flexible loop deletion by circular permutation in the streptavidin-biotin system.
Protein Sci., 7, 1998
|
|
1SWF
 
 | | CIRCULAR PERMUTED STREPTAVIDIN E51/A46 | | Descriptor: | CIRCULARLY PERMUTED CORE-STREPTAVIDIN E51/A46 | | Authors: | Freitag, S, Chu, V, Le Trong, I, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1997-04-23 | | Release date: | 1998-04-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Thermodynamic and structural consequences of flexible loop deletion by circular permutation in the streptavidin-biotin system.
Protein Sci., 7, 1998
|
|
5XOY
 
 | | Crystal structure of LysK from Thermus thermophilus in complex with Lysine | | Descriptor: | LYSINE, SULFATE ION, [LysW]-lysine hydrolase | | Authors: | Tomita, T, Fujita, S, Hasebe, F, Cho, S.-H, Yoshida, A, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2017-05-31 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystal structure of LysK, an enzyme catalyzing the last step of lysine biosynthesis in Thermus thermophilus, in complex with lysine: Insight into the mechanism for recognition of the amino-group carrier protein, LysW
Biochem. Biophys. Res. Commun., 491, 2017
|
|
9EUO
 
 | | Outward-open structure of Drosophila dopamine transporter bound to an atypical non-competitive inhibitor | | Descriptor: | 9D5 ANTIBODY, HEAVY CHAIN, LIGHT CHAIN, ... | | Authors: | Pedersen, C.N, Yang, F, Ita, S, Xu, Y, Akunuri, R, Trampari, S, Neumann, C.M.T, Desdorf, L.M, Schioett, B, Salvino, J.M, Mortensen, O.V, Nissen, P, Shahsavar, A. | | Deposit date: | 2024-03-27 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of the dopamine transporter with a novel atypical non-competitive inhibitor bound to the orthosteric site.
J.Neurochem., 168, 2024
|
|
9EUP
 
 | | Inhibitor-free outward-open structure of Drosophila dopamine transporter | | Descriptor: | 9D5 ANTIBODY, HEAVY CHAIN, LIGHT CHAIN, ... | | Authors: | Pedersen, C.N, Yang, F, Ita, S, Xu, Y, Akunuri, R, Trampari, S, Neumann, C.M.T, Desdorf, L.M, Schioett, B, Salvino, J.M, Mortensen, O.V, Nissen, P, Shahsavar, A. | | Deposit date: | 2024-03-27 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structure of the dopamine transporter with a novel atypical non-competitive inhibitor bound to the orthosteric site.
J.Neurochem., 168, 2024
|
|
4W96
 
 | | Crystal structure of cross-linked tetragonal hen egg white lysozyme soaked with 5mM [Ru(CO)3Cl2]2 followed by the reaction in deoxy-myoglobin solution | | Descriptor: | CHLORIDE ION, DIMETHYLFORMAMIDE, Lysozyme C, ... | | Authors: | Tabe, H, Fujita, K, Abe, S, Tsujimoto, M, Kuchimaru, T, Kizaka-Kondo, S, Takano, M, Kitagawa, S, Ueno, T. | | Deposit date: | 2014-08-27 | | Release date: | 2014-12-31 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Preparation of a Cross-Linked Porous Protein Crystal Containing Ru Carbonyl Complexes as a CO-Releasing Extracellular Scaffold
Inorg.Chem., 54, 2015
|
|
4W94
 
 | | Crystal structure of cross-linked tetragonal hen egg white lysozyme soaked with 5mM [Ru(CO)3Cl2]2 | | Descriptor: | CHLORIDE ION, DIMETHYLFORMAMIDE, Lysozyme C, ... | | Authors: | Tabe, H, Fujita, K, Abe, S, Tsujimoto, M, Kuchimaru, T, Kizaka-Kondo, S, Takano, M, Kitagawa, S, Ueno, T. | | Deposit date: | 2014-08-27 | | Release date: | 2014-12-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Preparation of a Cross-Linked Porous Protein Crystal Containing Ru Carbonyl Complexes as a CO-Releasing Extracellular Scaffold
Inorg.Chem., 54, 2015
|
|
8YIB
 
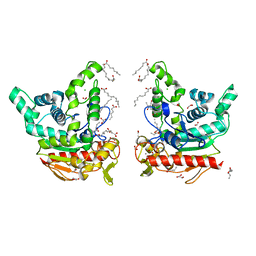 | | Staphylococcus aureus lipase -PSA complex - covalent bonding state | | Descriptor: | ACETIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kitadokoro, J, Kamitani, S, Kitadokoro, K. | | Deposit date: | 2024-02-29 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of Staphylococcus aureus lipase complex with unsaturated petroselinic acid.
Febs Open Bio, 14, 2024
|
|
2PSM
 
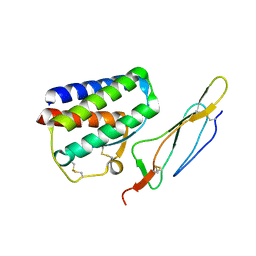 | | Crystal structure of Interleukin 15 in complex with Interleukin 15 receptor alpha | | Descriptor: | BENZAMIDINE, Interleukin-15, Interleukin-15 receptor alpha chain | | Authors: | Olsen, S.K, Murayama, K, Kishishita, S, Kukimoto-Niino, M, Terada, T, Shirouzu, M, Ota, N, Kanagawa, O, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-07 | | Release date: | 2007-11-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal Structure of the Interleukin-15{middle dot}Interleukin-15 Receptor {alpha} Complex: INSIGHTS INTO TRANS AND CIS PRESENTATION
J.Biol.Chem., 282, 2007
|
|
5WS3
 
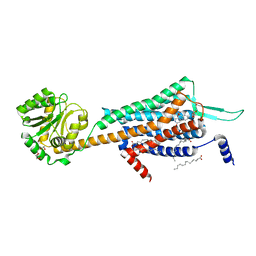 | | Crystal structures of human orexin 2 receptor bound to the selective antagonist EMPA determined by serial femtosecond crystallography at SACLA | | Descriptor: | N-ethyl-2-[(6-methoxypyridin-3-yl)-(2-methylphenyl)sulfonyl-amino]-N-(pyridin-3-ylmethyl)ethanamide, OLEIC ACID, Orexin receptor type 2,GlgA glycogen synthase,Orexin receptor type 2, ... | | Authors: | Suno, R, Kimura, K, Nakane, T, Yamashita, K, Wang, J, Fujiwara, T, Yamanaka, Y, Im, D, Tsujimoto, H, Sasanuma, M, Horita, S, Hirokawa, T, Nango, E, Tono, K, Kameshima, T, Hatsui, T, Joti, Y, Yabashi, M, Shimamoto, K, Yamamoto, M, Rosenbaum, D.M, Iwata, S, Shimamura, T, Kobayashi, T. | | Deposit date: | 2016-12-05 | | Release date: | 2017-12-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Human Orexin 2 Receptor Bound to the Subtype-Selective Antagonist EMPA.
Structure, 26, 2018
|
|
5WQC
 
 | | Crystal structure of human orexin 2 receptor bound to the selective antagonist EMPA determined by the synchrotron light source at SPring-8. | | Descriptor: | N-ethyl-2-[(6-methoxypyridin-3-yl)-(2-methylphenyl)sulfonyl-amino]-N-(pyridin-3-ylmethyl)ethanamide, OLEIC ACID, Orexin receptor type 2,GlgA glycogen synthase,Orexin receptor type 2, ... | | Authors: | Suno, R, Hirata, K, Yamashita, K, Tsujimoto, H, Sasanuma, M, Horita, S, Yamamoto, M, Rosenbaum, D.M, Iwata, S, Shimamura, T, Kobayashi, T. | | Deposit date: | 2016-11-25 | | Release date: | 2017-11-29 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal Structures of Human Orexin 2 Receptor Bound to the Subtype-Selective Antagonist EMPA
Structure, 26, 2018
|
|
8J9F
 
 | | Structure of STG-hydrolyzing beta-glucosidase 1 (PSTG1) | | Descriptor: | Beta-glucosidase, GLYCEROL | | Authors: | Yanai, T, Imaizumi, R, Takahashi, Y, Katsumura, E, Yamamoto, M, Nakayama, T, Yamashita, S, Takeshita, K, Sakai, N, Matsuura, H. | | Deposit date: | 2023-05-03 | | Release date: | 2024-04-10 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural insights into a bacterial beta-glucosidase capable of degrading sesaminol triglucoside to produce sesaminol: toward the understanding of the aglycone recognition mechanism by the C-terminal lid domain.
J.Biochem., 174, 2023
|
|
6LMI
 
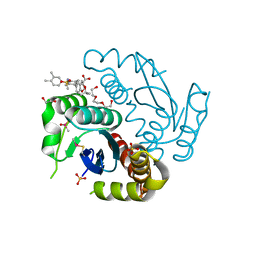 | | Crystal structure of HIV-1 integrase catalytic core domain in complex with 2-(tert-butoxy)-2-[3-(3,4-dihydro-2H-1-benzopyran-6-yl)-6-methanesulfonamido-2,3',4',5-tetramethyl-[1,1'-biphenyl]-4-yl]acetic acid | | Descriptor: | (2S)-2-[2-(3,4-dihydro-2H-chromen-6-yl)-4-(3,4-dimethylphenyl)-3,6-dimethyl-5-(methylsulfonylamino)phenyl]-2-[(2-methylpropan-2-yl)oxy]ethanoic acid, 1,2-ETHANEDIOL, Integrase catalytic, ... | | Authors: | Sugiyama, S, Iwaki, T, Tamura, Y, Tomita, K, Matsuoka, E, Arita, S, Seki, T, Yoshinaga, T, Kawasuji, T. | | Deposit date: | 2019-12-25 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of novel integrase-LEDGF/p75 allosteric inhibitors based on a benzene scaffold.
Bioorg.Med.Chem., 28, 2020
|
|
6LMQ
 
 | | Crystal structure of HIV-1 integrase catalytic core domain in complex with 2-(tert-butoxy)-2-[3-(3,4-dihydro-2H-1,4-benzoxazin-6-yl)-6-methanesulfonamido-2,3',4',5-tetramethyl-[1,1'-biphenyl]-4-yl]acetic acid | | Descriptor: | (2S)-2-[2-(3,4-dihydro-2H-1,4-benzoxazin-6-yl)-4-(3,4-dimethylphenyl)-3,6-dimethyl-5-(methylsulfonylamino)phenyl]-2-[(2-methylpropan-2-yl)oxy]ethanoic acid, Integrase catalytic, SULFATE ION, ... | | Authors: | Sugiyama, S, Iwaki, T, Tamura, Y, Tomita, K, Matsuoka, E, Arita, S, Seki, T, Yoshinaga, T, Kawasuji, T. | | Deposit date: | 2019-12-26 | | Release date: | 2020-09-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of novel integrase-LEDGF/p75 allosteric inhibitors based on a benzene scaffold.
Bioorg.Med.Chem., 28, 2020
|
|
