6C3K
 
 | |
6C3A
 
 | | O2-, PLP-dependent L-arginine hydroxylase RohP 4-hydroxy-2-ketoarginine complex | | Descriptor: | (4S)-5-carbamimidamido-4-hydroxy-2-oxopentanoic acid, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hedges, J.B, Ryan, K.S. | | Deposit date: | 2018-01-09 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Snapshots of the Catalytic Cycle of an O2, Pyridoxal Phosphate-Dependent Hydroxylase.
ACS Chem. Biol., 13, 2018
|
|
6C3C
 
 | | PLP-dependent L-arginine hydroxylase RohP quinonoid I complex | | Descriptor: | (2E)-5-carbamimidamido-2-{[(Z)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4(1H)-ylidene}methyl]imino}pentanoic acid, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hedges, J.B, Ryan, K.S. | | Deposit date: | 2018-01-09 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Snapshots of the Catalytic Cycle of an O2, Pyridoxal Phosphate-Dependent Hydroxylase.
ACS Chem. Biol., 13, 2018
|
|
2E86
 
 | |
2EI2
 
 | | Crystal Structure Analysis of the 1,2-dihydroxynaphthalene dioxygenase from Pseudomonas sp. stain C18 | | Descriptor: | 1,2-dihydroxynaphthalene dioxygenase, FE (II) ION, GLYCEROL, ... | | Authors: | Neau, D.B, Kelker, M.S, Colbert, C.L, Bolin, J.T. | | Deposit date: | 2007-03-10 | | Release date: | 2008-02-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural explanation for success and failure in the enzymatic ring-cleavage of 3,4 dihydroxybiphenyl and related PCB metabolites
To be Published
|
|
2EI1
 
 | | Anaerobic Crystal Structure Analysis of the 1,2-dihydroxynaphthalene dioxygeanse of Pseudomonas sp. strain C18 complexes to 1,2-dihydroxynaphthalene | | Descriptor: | 1,2-dihydroxynaphthalene dioxygenase, FE (II) ION, GLYCEROL, ... | | Authors: | Neau, D.B, Kelker, M.S, Colbert, C.L, Bolin, J.T. | | Deposit date: | 2007-03-10 | | Release date: | 2008-02-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structural explanation for success and failure in the enzymatic ring-cleavage of 3,4 dihydroxybiphenyl and related PCB metabolites.
To be Published
|
|
2EHZ
 
 | | Anaerobic Crystal Structure Analysis of 1,2-dihydroxynaphthalene dioxygenase from Pseudomonas sp. strain C18 complexed with 4-methylcatechol | | Descriptor: | 1,2-ETHANEDIOL, 1,2-dihydroxynaphthalene dioxygenase, 4-METHYLCATECHOL, ... | | Authors: | Neau, D.B, Kelker, M.S, Colbert, C.L, Bolin, J.T. | | Deposit date: | 2007-03-10 | | Release date: | 2008-02-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural explanation for success and failure in the enzymatic ring-cleavage of 3,4 dihydroxybiphenyl and related PCB metabolites
To be Published
|
|
2EI3
 
 | | Anaerobic Crystal Structure Analysis of the 1,2-dihydroxynaphthalene dioxygenase from Pseudomonas sp. strain C18 complexes with 2,3-dihydroxybiphenyl | | Descriptor: | 1,2-dihydroxynaphthalene dioxygenase, BIPHENYL-2,3-DIOL, FE (II) ION, ... | | Authors: | Neau, D.B, Kelker, M.S, Colbert, C.L, Bolin, J.T. | | Deposit date: | 2007-03-10 | | Release date: | 2008-02-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural explanation for success and failure in the enzymatic ring-cleavage of 3,4 dihydroxybiphenyl and related PCB metabolites
To be Published
|
|
2EI0
 
 | | Anaerobic Crystal Structure Analysis of 1,2-dihydroxynaphthalene dioxygenase from Pseudomonas sp. strain C18 complexed with 3,4-dihydroxybiphenyl | | Descriptor: | 1,1'-BIPHENYL-3,4-DIOL, 1,2-dihydroxynaphthalene dioxygenase, FE (II) ION, ... | | Authors: | Neau, D.B, Kelker, M.S, Colbert, C.L, Bolin, J.T. | | Deposit date: | 2007-03-10 | | Release date: | 2008-02-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural explanation for success and failure in the enzymatic ring-cleavage of 3,4 dihydroxybiphenyl and related PCB metabolites
To be Published
|
|
5TW4
 
 | |
5TY7
 
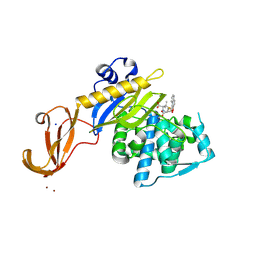 | | Crystal structure of wild-type S. aureus penicillin binding protein 4 (PBP4) in complex with nafcillin | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2-ethoxynaphthalen-1-yl)carbonyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Penicillin-binding protein 4, SODIUM ION, ... | | Authors: | Alexander, J.A.N, Strynadka, N.C.J. | | Deposit date: | 2016-11-18 | | Release date: | 2018-06-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.894 Å) | | Cite: | Structural and kinetic analysis of penicillin-binding protein 4 (PBP4)-mediated antibiotic resistance inStaphylococcus aureus.
J. Biol. Chem., 2018
|
|
5TX9
 
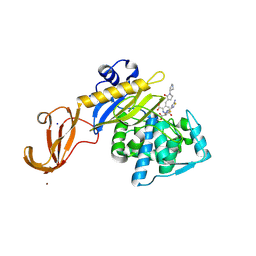 | | Crystal structure of S. aureus penicillin binding protein 4 (PBP4) mutant (E183A, F241R) in complex with ceftobiprole | | Descriptor: | (2R)-2-[(1R)-1-{[(2Z)-2-(5-amino-1,2,4-thiadiazol-3-yl)-2-(hydroxyimino)acetyl]amino}-2-oxoethyl]-5-({2-oxo-1-[(3R)-pyr rolidin-3-yl]-2,5-dihydro-1H-pyrrol-3-yl}methyl)-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Penicillin-binding protein 4, SODIUM ION, ... | | Authors: | Alexander, J.A.N, Strynadka, N.C.J. | | Deposit date: | 2016-11-16 | | Release date: | 2018-05-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural and kinetic analysis of penicillin-binding protein 4 (PBP4)-mediated antibiotic resistance inStaphylococcus aureus.
J. Biol. Chem., 2018
|
|
5TW8
 
 | |
5TXI
 
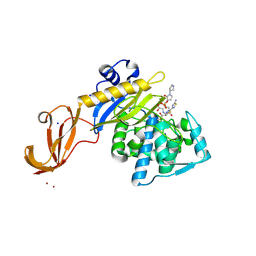 | | Crystal structure of wild-type S. aureus penicillin binding protein 4 (PBP4) in complex with ceftobiprole | | Descriptor: | (2R)-2-[(1R)-1-{[(2Z)-2-(5-amino-1,2,4-thiadiazol-3-yl)-2-(hydroxyimino)acetyl]amino}-2-oxoethyl]-5-({2-oxo-1-[(3R)-pyr rolidin-3-yl]-2,5-dihydro-1H-pyrrol-3-yl}methyl)-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Alexander, J.A.N, Strynadka, N.C.J. | | Deposit date: | 2016-11-16 | | Release date: | 2018-05-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and kinetic analysis of penicillin-binding protein 4 (PBP4)-mediated antibiotic resistance inStaphylococcus aureus.
J. Biol. Chem., 2018
|
|
3V1N
 
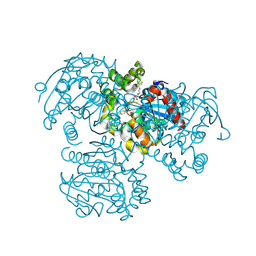 | | Crystal Structure of the H265Q mutant of a C-C hydrolase, BphD from Burkholderia xenovorans LB400, after exposure to its substrate HOPDA | | Descriptor: | (3E)-2,6-DIOXO-6-PHENYLHEX-3-ENOATE, 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate hydrolase, BENZOIC ACID, ... | | Authors: | Ghosh, S, Bolin, J.T. | | Deposit date: | 2011-12-09 | | Release date: | 2012-03-21 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Identification of an Acyl-Enzyme Intermediate in a meta-Cleavage Product Hydrolase Reveals the Versatility of the Catalytic Triad.
J.Am.Chem.Soc., 134, 2012
|
|
3V1K
 
 | |
3V1L
 
 | |
5VMM
 
 | | Staphylococcus aureus IsdB bound to human hemoglobin | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, Iron-regulated cell wall-anchored protein, ... | | Authors: | Bowden, C.F, Chan, A.C, Murphy, M.E. | | Deposit date: | 2017-04-27 | | Release date: | 2017-11-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure-function analyses reveal key features in Staphylococcus aureus IsdB-associated unfolding of the heme-binding pocket of human hemoglobin.
J. Biol. Chem., 293, 2018
|
|
3V1M
 
 | | Crystal Structure of the S112A/H265Q mutant of a C-C hydrolase, BphD from Burkholderia xenovorans LB400, after exposure to its substrate HOPDA | | Descriptor: | (3E)-2,6-DIOXO-6-PHENYLHEX-3-ENOATE, 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate hydrolase, MALONATE ION | | Authors: | Ghosh, S, Bolin, J.T. | | Deposit date: | 2011-12-09 | | Release date: | 2012-03-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Identification of an Acyl-Enzyme Intermediate in a meta-Cleavage Product Hydrolase Reveals the Versatility of the Catalytic Triad.
J.Am.Chem.Soc., 134, 2012
|
|
5TY2
 
 | | Crystal structure of S. aureus penicillin binding protein 4 (PBP4) mutant (E183A, F241R) in complex with nafcillin | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2-ethoxynaphthalen-1-yl)carbonyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, CHLORIDE ION, Penicillin-binding protein 4, ... | | Authors: | Alexander, J.A.N, Strynadka, N.C.J. | | Deposit date: | 2016-11-18 | | Release date: | 2018-06-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and kinetic analysis of penicillin-binding protein 4 (PBP4)-mediated antibiotic resistance inStaphylococcus aureus.
J. Biol. Chem., 2018
|
|
3VB0
 
 | | Crystal structure of 2,2',3-trihydroxybiphenyl 1,2-dioxygenase from dibenzofuran-degrading Sphingomonas wittichii strain RW1 | | Descriptor: | FE (II) ION, Glyoxalase/bleomycin resistance protein/dioxygenase, HEXAETHYLENE GLYCOL, ... | | Authors: | Koksal, M, Kumar, P, Bolin, J.T. | | Deposit date: | 2011-12-30 | | Release date: | 2013-04-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a dibenzofuran-degrading dioxygenase: an unusual spatially heterogeneous crystal with a hypersymmetric intensity distribution
To be Published
|
|
2OG1
 
 | | Crystal Structure of BphD, a C-C hydrolase from Burkholderia xenovorans LB400 | | Descriptor: | 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate hydrolase, ETHANOL, GLYCEROL, ... | | Authors: | Dai, S, Ke, J, Bolin, J.T. | | Deposit date: | 2007-01-04 | | Release date: | 2007-01-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Kinetic and structural insight into the mechanism of BphD, a C-C bond hydrolase from the biphenyl degradation pathway
Biochemistry, 45, 2006
|
|
2PP9
 
 | | Nitrate bound wild type oxidized AfNiR | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, COPPER (I) ION, ... | | Authors: | Murphy, M.E.P, Tocheva, E.I. | | Deposit date: | 2007-04-28 | | Release date: | 2008-04-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conserved active site residues limit inhibition of a copper-containing nitrite reductase by small molecules.
Biochemistry, 47, 2008
|
|
2PP7
 
 | |
2PPA
 
 | |
