6OVT
 
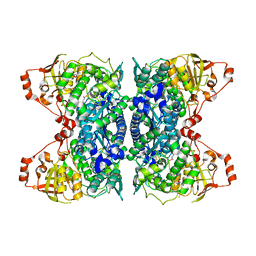 | | Crystal Structure of IlvD from Mycobacterium tuberculosis | | Descriptor: | DI(HYDROXYETHYL)ETHER, Dihydroxy-acid dehydratase, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Almo, S.C, Grove, T.L, Bonanno, J.B, Baker, E.N, Bashiri, G. | | Deposit date: | 2019-05-08 | | Release date: | 2019-08-07 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The active site of theMycobacterium tuberculosisbranched-chain amino acid biosynthesis enzyme dihydroxyacid dehydratase contains a 2Fe-2S cluster.
J.Biol.Chem., 294, 2019
|
|
4JLF
 
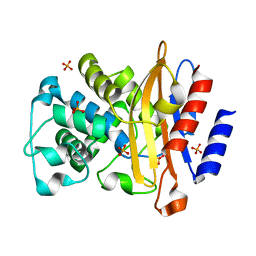 | | Inhibitor resistant (R220A) substitution in the Mycobacterium tuberculosis beta-lactamase | | Descriptor: | Beta-lactamase, PHOSPHATE ION | | Authors: | Hazra, S, Kurz, S, Blanchard, J, Bonomo, R. | | Deposit date: | 2013-03-12 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Can inhibitor-resistant substitutions in the Mycobacterium tuberculosis beta-Lactamase BlaC lead to clavulanate resistance?: a biochemical rationale for the use of beta-lactam-beta-lactamase inhibitor combinations.
Antimicrob.Agents Chemother., 57, 2013
|
|
4M0D
 
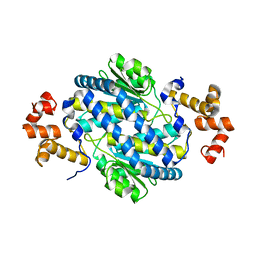 | |
4LZJ
 
 | | Crystal Structure of MurQ from H.influenzae with bound inhibitor | | Descriptor: | 2-(acetylamino)-3-O-[(1R)-1-carboxyethyl]-2-deoxy-6-O-phosphono-D-glucitol, N-acetylmuramic acid 6-phosphate etherase, PHOSPHATE ION | | Authors: | Hazra, S, Blanchard, J. | | Deposit date: | 2013-07-31 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.405 Å) | | Cite: | Structure of MurNAc 6-phosphate hydrolase (MurQ) from Haemophilus influenzae with a bound inhibitor.
Biochemistry, 52, 2013
|
|
1C1X
 
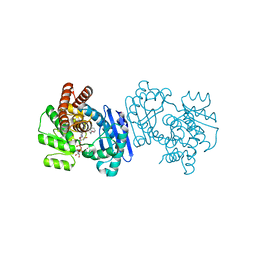 | | L-PHENYLALANINE DEHYDROGENASE STRUCTURE IN TERNARY COMPLEX WITH NAD+ AND L-3-PHENYLLACTATE | | Descriptor: | ALPHA-HYDROXY-BETA-PHENYL-PROPIONIC ACID, ISOPROPYL ALCOHOL, L-PHENYLALANINE DEHYDROGENASE, ... | | Authors: | Vanhooke, J.L, Thoden, J.B. | | Deposit date: | 1999-07-22 | | Release date: | 2000-08-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Rhodococcus L-phenylalanine dehydrogenase: kinetics, mechanism, and structural basis for catalytic specificity.
Biochemistry, 39, 2000
|
|
1C1D
 
 | | L-PHENYLALANINE DEHYDROGENASE STRUCTURE IN TERNARY COMPLEX WITH NADH AND L-PHENYLALANINE | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ISOPROPYL ALCOHOL, L-PHENYLALANINE DEHYDROGENASE, ... | | Authors: | Vanhooke, J.L, Thoden, J.B. | | Deposit date: | 1999-07-21 | | Release date: | 2000-08-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Rhodococcus L-phenylalanine dehydrogenase: kinetics, mechanism, and structural basis for catalytic specificity.
Biochemistry, 39, 2000
|
|
1ZID
 
 | |
