2RJ1
 
 | |
2RIT
 
 | | Unliganded B-specific-1,3-galactosyltransferase (GTB) | | Descriptor: | GLYCEROL, Glycoprotein-fucosylgalactoside alpha-galactosyltransferase | | Authors: | Evans, S.V, Alfaro, J.A. | | Deposit date: | 2007-10-12 | | Release date: | 2008-02-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | ABO(H) blood group A and B glycosyltransferases recognize substrate via specific conformational changes.
J.Biol.Chem., 283, 2008
|
|
2RJ7
 
 | |
2RJ6
 
 | |
2RIZ
 
 | |
2RJ5
 
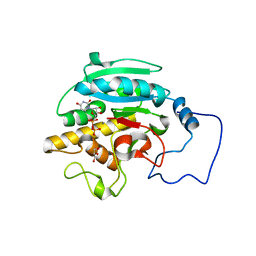 | |
5L1Z
 
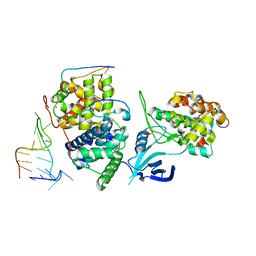 | | TAR complex with HIV-1 Tat-AFF4-P-TEFb | | Descriptor: | AF4/FMR2 family member 4, Cyclin-T1, Cyclin-dependent kinase 9, ... | | Authors: | Schulze-Gahmen, U, Hurley, J. | | Deposit date: | 2016-07-29 | | Release date: | 2016-10-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (5.9 Å) | | Cite: | Insights into HIV-1 proviral transcription from integrative structure and dynamics of the Tat:AFF4:P-TEFb:TAR complex.
Elife, 5, 2016
|
|
3RDH
 
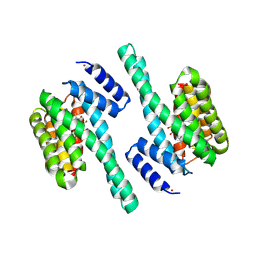 | | X-ray induced covalent inhibition of 14-3-3 | | Descriptor: | 14-3-3 protein zeta/delta, 4-[(E)-{4-formyl-5-hydroxy-6-methyl-3-[(phosphonooxy)methyl]pyridin-2-yl}diazenyl]benzoic acid, NICKEL (II) ION | | Authors: | Horton, J.R, Upadhyay, A.K, Fu, H, Cheng, X. | | Deposit date: | 2011-04-01 | | Release date: | 2011-09-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Discovery and structural characterization of a small molecule 14-3-3 protein-protein interaction inhibitor.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
8XJV
 
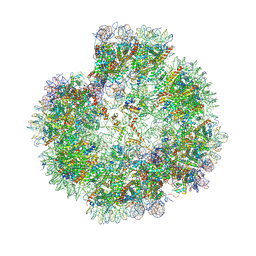 | | Structural basis for the linker histone H5-nucleosome binding and chromatin compaction | | Descriptor: | DNA, Histone H2A, Histone H2B 1.1, ... | | Authors: | Li, W.Y, Song, F, Zhu, P. | | Deposit date: | 2023-12-22 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for linker histone H5-nucleosome binding and chromatin fiber compaction.
Cell Res., 34, 2024
|
|
6PWF
 
 | |
6PWE
 
 | | Cryo-EM structure of nucleosome core particle | | Descriptor: | DNA (147-MER), Histone H2A, Histone H2B, ... | | Authors: | Chittori, S, Subramaniam, S. | | Deposit date: | 2019-07-22 | | Release date: | 2019-08-21 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Structure of the primed state of the ATPase domain of chromatin remodeling factor ISWI bound to the nucleosome.
Nucleic Acids Res., 47, 2019
|
|
2PCX
 
 | | Crystal structure of p53DBD(R282Q) at 1.54-angstrom Resolution | | Descriptor: | Cellular tumor antigen p53, ZINC ION | | Authors: | Tu, C, Shaw, G, Ji, X. | | Deposit date: | 2007-03-30 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Impact of low-frequency hotspot mutation R282Q on the structure of p53 DNA-binding domain as revealed by crystallography at 1.54 angstroms resolution.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
6E0P
 
 | |
6DZT
 
 | |
6E0C
 
 | |
5WCU
 
 | |
3PX8
 
 | | RTA in complex with 7-carboxy-pterin | | Descriptor: | 2-amino-4-oxo-1,4-dihydropteridine-7-carboxylic acid, Preproricin | | Authors: | Jasheway, K.R, Robertus, J.D. | | Deposit date: | 2010-12-09 | | Release date: | 2011-06-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | 7-Substituted pterins provide a new direction for ricin A chain inhibitors.
Eur.J.Med.Chem., 46, 2011
|
|
3PX9
 
 | |
7E7Y
 
 | |
7E8C
 
 | | SARS-CoV-2 S-6P in complex with 9 Fabs | | Descriptor: | 368-2 H, 368-2 L, 604 H, ... | | Authors: | Du, S, Xiao, J, Zhang, Z. | | Deposit date: | 2021-03-01 | | Release date: | 2021-06-09 | | Last modified: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Humoral immune response to circulating SARS-CoV-2 variants elicited by inactivated and RBD-subunit vaccines.
Cell Res., 31, 2021
|
|
7E7X
 
 | |
7E88
 
 | |
7E8F
 
 | | SARS-CoV-2 NTD in complex with N9 Fab | | Descriptor: | 368-2 H, 368-2 L, 604 H, ... | | Authors: | Du, S, Xiao, J, Zhang, Z. | | Deposit date: | 2021-03-01 | | Release date: | 2021-06-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Humoral immune response to circulating SARS-CoV-2 variants elicited by inactivated and RBD-subunit vaccines.
Cell Res., 31, 2021
|
|
7E86
 
 | |
6O1D
 
 | |
