2EB9
 
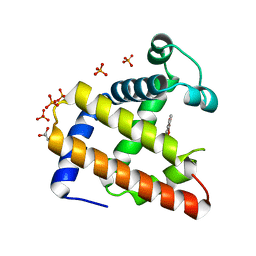 | | Crystal Structure of Cu(II)(Sal-Leu)/apo-Myoglobin | | Descriptor: | (N-SALICYLIDEN-L-LEUCINATO)-COPPER(II), GLYCEROL, Myoglobin, ... | | Authors: | Abe, S, Okazaki, S, Ueno, T, Hikage, T, Suzuki, A, Yamane, T, Watanabe, Y. | | Deposit date: | 2007-02-07 | | Release date: | 2007-07-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design and Structure Analysis of Artificial Metalloproteins: Selective Coordination of His64 to Copper Complexes with Square-Planar Structure in the apo-Myoglobin Scaffold
Inorg.Chem., 46, 2007
|
|
4K9G
 
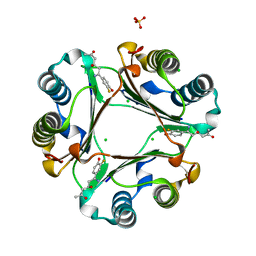 | | 1.55 A Crystal Structure of Macrophage Migration Inhibitory Factor bound to ISO-66 and a related compound | | Descriptor: | (4R,6Z)-6-(3-fluoro-4-hydroxyphenyl)-4-hydroxy-6-iminohexan-2-one, 1-[(5S)-3-(3-fluoro-4-hydroxyphenyl)-4,5-dihydro-1,2-oxazol-5-yl]propan-2-one, CHLORIDE ION, ... | | Authors: | Crichlow, G.V, Al-Abed, Y, Lolis, E.J. | | Deposit date: | 2013-04-19 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | ISO-66, a novel inhibitor of macrophage migration, shows efficacy in melanoma and colon cancer models.
INT J ONCOL., 45, 2014
|
|
4WCO
 
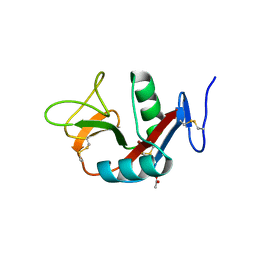 | | Crystal structure of extracellular domain of human lectin-like transcript 1 (LLT1), the ligand for natural killer receptor-P1A | | Descriptor: | ACETATE ION, C-type lectin domain family 2 member D, SULFATE ION, ... | | Authors: | Kita, S, Matsubara, H, Kasai, Y, Tamaoki, T, Okabe, Y, Fukuhara, H, Kamishikiryo, J, Ose, T, Kuroki, K, Maenaka, K. | | Deposit date: | 2014-09-05 | | Release date: | 2015-06-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Crystal structure of extracellular domain of human lectin-like transcript 1 (LLT1), the ligand for natural killer receptor-P1A
Eur.J.Immunol., 45, 2015
|
|
1LJT
 
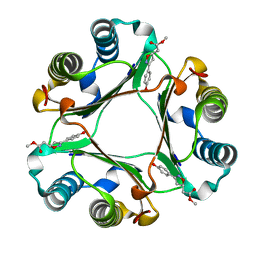 | | Crystal Structure of Macrophage Migration Inhibitory Factor complexed with (S,R)-3-(4-hydroxyphenyl)-4,5-dihydro-5-isoxazole-acetic acid methyl ester (ISO-1) | | Descriptor: | 3-(4-HYDROXYPHENYL)-4,5-DIHYDRO-5-ISOXAZOLE-ACETIC ACID METHYL ESTER, Macrophage migration inhibitory factor | | Authors: | Lubetsky, J.B, Dios, A, Han, J, Aljabari, B, Ruzsicska, B, Mitchell, R, Lolis, E, Al-Abed, Y. | | Deposit date: | 2002-04-22 | | Release date: | 2002-11-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Tautomerase Active Site of Macrophage Migration Inhibitory factor is a Potential Target for Discovery of Novel Anti-inflammatory Agents
J.Biol.Chem., 277, 2002
|
|
2OOH
 
 | | Crystal Structure of MIF bound to a Novel Inhibitor, OXIM-11 | | Descriptor: | 4-HYDROXYBENZALDEHYDE O-(CYCLOHEXYLCARBONYL)OXIME, GLYCEROL, Macrophage migration inhibitory factor, ... | | Authors: | Crichlow, G.V, Al-Abed, Y, Lolis, E. | | Deposit date: | 2007-01-25 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Alternative chemical modifications reverse the binding orientation of a pharmacophore scaffold in the active site of macrophage migration inhibitory factor.
J.Biol.Chem., 282, 2007
|
|
2OOW
 
 | | MIF Bound to a Fluorinated OXIM Derivative | | Descriptor: | 3-FLUORO-4-HYDROXYBENZALDEHYDE O-(CYCLOHEXYLCARBONYL)OXIME, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Crichlow, G.V, Al-Abed, Y, Lolis, E. | | Deposit date: | 2007-01-26 | | Release date: | 2007-06-05 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Alternative chemical modifications reverse the binding orientation of a pharmacophore scaffold in the active site of macrophage migration inhibitory factor.
J.Biol.Chem., 282, 2007
|
|
2OOZ
 
 | | Macrophage Migration Inhibitory Factor (MIF) Complexed with OXIM6 (an OXIM Derivative Not Containing a Ring in its R-group) | | Descriptor: | 4-HYDROXYBENZALDEHYDE O-(3,3-DIMETHYLBUTANOYL)OXIME, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Crichlow, G.V, Al-Abed, Y, Lolis, E. | | Deposit date: | 2007-01-26 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Alternative chemical modifications reverse the binding orientation of a pharmacophore scaffold in the active site of macrophage migration inhibitory factor.
J.Biol.Chem., 282, 2007
|
|
2DQA
 
 | | Crystal Structure of Tapes japonica Lysozyme | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lysozyme, PLATINUM (II) ION, ... | | Authors: | Goto, T, Kakuta, Y, Abe, Y, Takeshita, K, Imoto, T, Ueda, T. | | Deposit date: | 2006-05-24 | | Release date: | 2007-06-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Tapes japonica Lysozyme with Substrate Analogue: STRUCTURAL BASIS OF THE CATALYTIC MECHANISM AND MANIFESTATION OF ITS CHITINASE ACTIVITY ACCOMPANIED BY QUATERNARY STRUCTURAL CHANGE
J.Biol.Chem., 282, 2007
|
|
4LMU
 
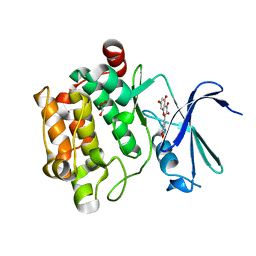 | | Crystal structure of Pim1 in complex with the inhibitor Quercetin (resulting from displacement of SKF86002) | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, GLYCEROL, Serine/threonine-protein kinase pim-1 | | Authors: | Parker, L.J, Tanaka, A, Handa, N, Honda, K, Tomabechi, Y, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2013-07-11 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Kinase crystal identification and ATP-competitive inhibitor screening using the fluorescent ligand SKF86002.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4LL5
 
 | | Crystal Structure of Pim-1 in complex with the fluorescent compound SKF86002 | | Descriptor: | 6-(4-fluorophenyl)-5-(pyridin-4-yl)-2,3-dihydroimidazo[2,1-b][1,3]thiazole, CALCIUM ION, GLYCEROL, ... | | Authors: | Parker, L.J, Tanaka, A, Handa, N, Honda, K, Tomabechi, Y, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2013-07-09 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kinase crystal identification and ATP-competitive inhibitor screening using the fluorescent ligand SKF86002.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5XHL
 
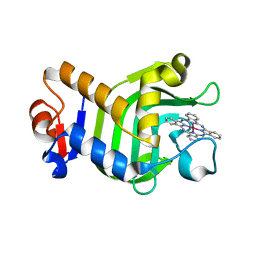 | | Crystal Structure of HasAp with Gallium Phthalocyanine | | Descriptor: | Heme acquisition protein HasAp, Phthalocyanine containing GA | | Authors: | Shoji, O, Shisaka, Y, Iwai, Y, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2017-04-21 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of HasAp with Gallium Phthalocyanine
to be published
|
|
5XIE
 
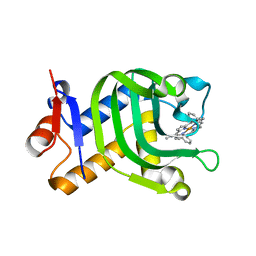 | | Crystal Structure of HasAp with 5-ethynyl-10,20-diphenylporphyrin | | Descriptor: | 5-Ethynyl-10,20-diphenylporphyrin containing FE, Heme acquisition protein HasAp | | Authors: | Shoji, O, Uehara, H, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2017-04-26 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structures of the Heme Acquisition Protein HasA with Iron(III)-5,15-Diphenylporphyrin and Derivatives Thereof as an Artificial Prosthetic Group
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5XHJ
 
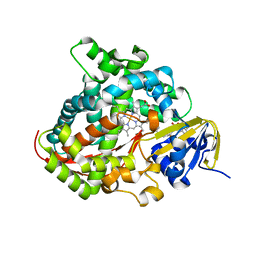 | | Crystal Structure of P450BM3 with 5-Cyclohexylvaleroyl-L-Tryptophan | | Descriptor: | 5-cyclohexylpentanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Suzuki, K, Shoji, O, Stanfield, J.K, Kasai, C, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2017-04-21 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Control of stereoselectivity of benzylic hydroxylation catalysed by wild-type cytochrome P450BM3 using decoy molecules
CATALYSIS SCIENCE AND TECHNOLOGY, 7, 2017
|
|
7FGP
 
 | |
3A5P
 
 | | Crystal structure of hemagglutinin | | Descriptor: | Haemagglutinin I | | Authors: | Watanabe, N, Sakai, N, Nakamura, T, Nabeshima, Y, Kouno, T, Mizuguchi, M, Kawano, K. | | Deposit date: | 2009-08-10 | | Release date: | 2010-08-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The Structure of Physarum polycephalum hemagglutinin I suggests a minimal carbohydrate recognition domain of legume lectin fold
J.Mol.Biol., 405, 2011
|
|
1UFP
 
 | | Crystal Structure of an Artificial Metalloprotein:Fe(III)(3,3'-Me2-salophen)/apo-wild type Myoglobin | | Descriptor: | Myoglobin, PHOSPHATE ION | | Authors: | Ueno, T, Ohashi, M, Kono, M, Kondo, K, Suzuki, A, Yamane, T, Watanabe, Y. | | Deposit date: | 2003-06-04 | | Release date: | 2004-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Artificial Metalloproteins: Tight Binding of Fe(III)(Schiff-Base) by Mutation of Ala71 to Gly in Apo-Myoglobin
Inorg.Chem., 43, 2004
|
|
1UF2
 
 | | The Atomic Structure of Rice dwarf Virus (RDV) | | Descriptor: | Core protein P3, Outer capsid protein P8, Structural protein P7 | | Authors: | Nakagawa, A, Miyazaki, N, Taka, J, Naitow, H, Ogawa, A, Fujimoto, Z, Mizuno, H, Higashi, T, Watanabe, Y, Omura, T, Cheng, R.H, Tsukihara, T. | | Deposit date: | 2003-05-23 | | Release date: | 2003-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The atomic structure of rice dwarf virus reveals the self-assembly mechanism of component proteins.
Structure, 11, 2003
|
|
1UFJ
 
 | | Crystal Structure of an Artificial Metalloprotein:Fe(III)(3,3'-Me2-salophen)/apo-A71G Myoglobin | | Descriptor: | 'N,N'-BIS-(2-HYDROXY-3-METHYL-BENZYLIDENE)-BENZENE-1,2-DIAMINE', FE (III) ION, MYOGLOBIN, ... | | Authors: | Ueno, T, Ohashi, M, Kono, M, Kondo, K, Suzuki, A, Yamane, T, Watanabe, Y. | | Deposit date: | 2003-05-30 | | Release date: | 2004-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of Artificial Metalloproteins: Tight Binding of Fe(III)(Schiff-Base) by Mutation of Ala71 to Gly in Apo-Myoglobin
Inorg.Chem., 43, 2004
|
|
5JR8
 
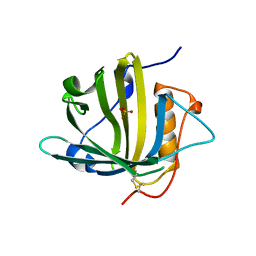 | | Disposal of Iron by a Mutant form of Siderocalin NGAL | | Descriptor: | GLYCEROL, Neutrophil gelatinase-associated lipocalin, PHOSPHATE ION | | Authors: | Rupert, P.B, Strong, R.K, Barasch, J, Hollman, M, Deng, R, Hod, E.A, Abergel, R, Allred, B, Xu, K, Darrah, S, Tekabe, Y, Perlstein, A, Bruck, E, Stauber, J, Corbin, K, Buchen, C, Slavkovich, V, Graziano, J, Spitalnik, S, Qiu, A. | | Deposit date: | 2016-05-05 | | Release date: | 2016-09-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Disposal of iron by a mutant form of lipocalin 2.
Nat Commun, 7, 2016
|
|
8I75
 
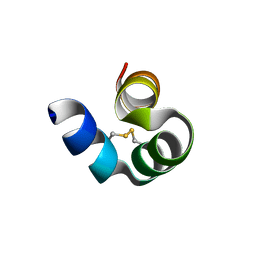 | |
8I76
 
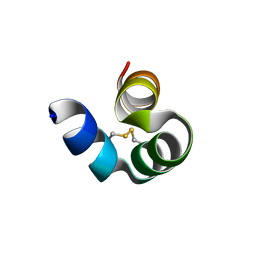 | |
8I74
 
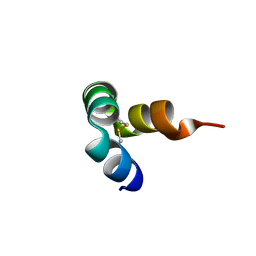 | |
5GHI
 
 | | Crystal structure of human MTH1(G2K mutant) in complex with 8-oxo-dGTP | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, SODIUM ION | | Authors: | Nakamura, T, Waz, S, Hirata, K, Nakabeppu, Y, Yamagata, Y. | | Deposit date: | 2016-06-20 | | Release date: | 2017-01-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.211 Å) | | Cite: | Structural and Kinetic Studies of the Human Nudix Hydrolase MTH1 Reveal the Mechanism for Its Broad Substrate Specificity
J. Biol. Chem., 292, 2017
|
|
5GHO
 
 | | Crystal structure of human MTH1(G2K/D120A mutant) in complex with 8-oxo-dGTP | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | Authors: | Nakamura, T, Waz, S, Hirata, K, Nakabeppu, Y, Yamagata, Y. | | Deposit date: | 2016-06-20 | | Release date: | 2017-01-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.191 Å) | | Cite: | Structural and Kinetic Studies of the Human Nudix Hydrolase MTH1 Reveal the Mechanism for Its Broad Substrate Specificity
J. Biol. Chem., 292, 2017
|
|
5GHN
 
 | | Crystal structure of human MTH1(G2K/D120N mutant) in complex with 2-oxo-dATP | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, SODIUM ION, [[(2R,3S,5R)-5-(6-azanyl-2-oxidanylidene-1H-purin-9-yl)-3-oxidanyl-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Nakamura, T, Waz, S, Hirata, K, Nakabeppu, Y, Yamagata, Y. | | Deposit date: | 2016-06-20 | | Release date: | 2017-01-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.391 Å) | | Cite: | Structural and Kinetic Studies of the Human Nudix Hydrolase MTH1 Reveal the Mechanism for Its Broad Substrate Specificity
J. Biol. Chem., 292, 2017
|
|
