7WN6
 
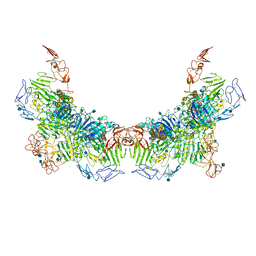 | | Cryo-EM structure of VWF D'D3 dimer (R1136M/E1143M mutant) complexed with D1D2 at 3.29 angstron resolution (2 units) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, von Willebrand antigen 2, ... | | Authors: | Zeng, J.W, Shu, Z.M, Zhou, A.W. | | Deposit date: | 2022-01-17 | | Release date: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structural mechanism of VWF D'D3 dimer formation.
Cell Discov, 8, 2022
|
|
7WN4
 
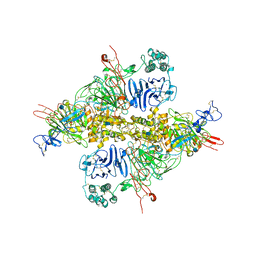 | | Cryo-EM structure of VWF D'D3 dimer (wild type) complexed with D1D2 at 3.4 angstron resolution (1 unit) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, von Willebrand antigen 2, ... | | Authors: | Zeng, J.W, Shu, Z.M, Zhou, A.W. | | Deposit date: | 2022-01-17 | | Release date: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural mechanism of VWF D'D3 dimer formation.
Cell Discov, 8, 2022
|
|
7WN3
 
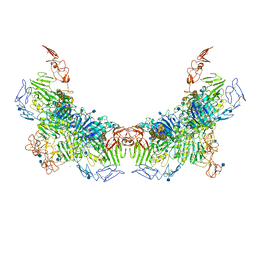 | | Cryo-EM structure of VWF D'D3 dimer (2M mutant) complexed with D1D2 at 3.29 angstron resolution (2 units) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, von Willebrand antigen 2, ... | | Authors: | Zeng, J.W, Shu, Z.M, Zhou, A.W. | | Deposit date: | 2022-01-17 | | Release date: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structural mechanism of VWF D'D3 dimer formation.
Cell Discov, 8, 2022
|
|
7X2A
 
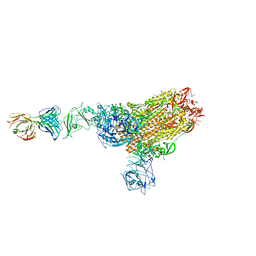 | | MERS-CoV spike complex with S41 neutralizing antibody Fab Class1 (1u2d RBD with 1Fab) | | Descriptor: | MERS-CoV Spike glycoprotein, antibody S41 heavy chain, antibody S41 light chain | | Authors: | Zeng, J.W, Zhang, S.Y, Zhou, H.X, Wang, X.W. | | Deposit date: | 2022-02-25 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Cryoelectron microscopy structures of a human neutralizing antibody bound to MERS-CoV spike glycoprotein.
Front Microbiol, 13, 2022
|
|
7X26
 
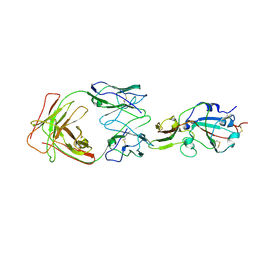 | | S41 neutralizing antibody Fab(MERS-CoV) | | Descriptor: | Spike glycoprotein, antibody S41 heavy chain, antibody S41 light chain | | Authors: | Zeng, J.W, Zhang, S.Y, Wang, X.W. | | Deposit date: | 2022-02-25 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.685 Å) | | Cite: | Cryoelectron microscopy structures of a human neutralizing antibody bound to MERS-CoV spike glycoprotein.
Front Microbiol, 13, 2022
|
|
7X29
 
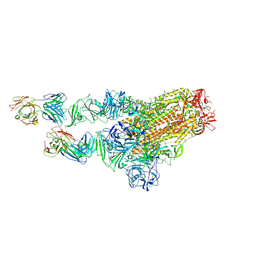 | | MERS-CoV spike complex with S41 neutralizing antibody Fab Class2 (1u2d RBD with 2Fab) | | Descriptor: | Spike glycoprotein, antibody S41 heavy chain, antibody S41 light chain | | Authors: | Zeng, J.W, Zhang, S.Y, Zhou, H.X, Wang, X.W. | | Deposit date: | 2022-02-25 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Cryoelectron microscopy structures of a human neutralizing antibody bound to MERS-CoV spike glycoprotein.
Front Microbiol, 13, 2022
|
|
7X28
 
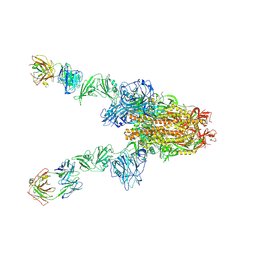 | | MERS-CoV spike complex with S41 neutralizing antibody Fab Class3 (2u1d RBD with 2Fab) | | Descriptor: | Spike glycoprotein, antibody S41 heavy chain, antibody S41 light chain | | Authors: | Zeng, J.W, Zhang, S.Y, Zhou, H.X, Wang, X.W. | | Deposit date: | 2022-02-25 | | Release date: | 2023-01-18 | | Last modified: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Cryoelectron microscopy structures of a human neutralizing antibody bound to MERS-CoV spike glycoprotein.
Front Microbiol, 13, 2022
|
|
5ZOV
 
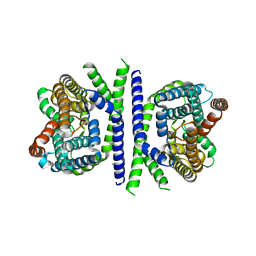 | | Inward-facing conformation of L-ascorbate transporter UlaA | | Descriptor: | ASCORBIC ACID, CALCIUM ION, PTS ascorbate-specific subunit IIBC | | Authors: | Wang, J.W. | | Deposit date: | 2018-04-16 | | Release date: | 2018-08-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.333 Å) | | Cite: | Inward-facing conformation of l-ascorbate transporter suggests an elevator mechanism
Cell Discov, 4, 2018
|
|
7X8X
 
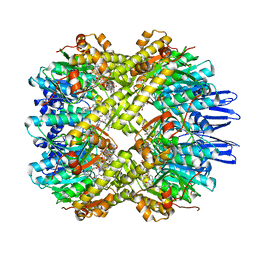 | | structural insights into Mycobacterium tuberculosis ClpP1P2 inhibition by Cediranib: implications for developing antimicrobial agents targeting Clp protease | | Descriptor: | 4-[1-[3-[4-[(4-fluoranyl-2-methyl-1H-indol-5-yl)oxy]-6-methoxy-quinazolin-7-yl]oxypropyl]piperidin-4-yl]benzamide, 4-[[3,5-bis(fluoranyl)phenyl]methyl]-N-[(4-bromophenyl)methyl]piperazine-1-carboxamide, ATP-dependent Clp protease proteolytic subunit 1, ... | | Authors: | Bao, R, Luo, Y.F, Zhu, Y.B, Yang, Y, Zhou, Y.Z. | | Deposit date: | 2022-03-15 | | Release date: | 2023-04-12 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Discovery and Mechanistic Study of Novel Mycobacterium tuberculosis ClpP1P2 Inhibitors
J.Med.Chem., 66, 2023
|
|
7VEH
 
 | | Type I-F Anti-CRISPR protein AcrIF13 | | Descriptor: | AcrIF13 | | Authors: | Gao, T, Feng, Y. | | Deposit date: | 2021-09-08 | | Release date: | 2022-07-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Mechanistic insights into the inhibition of the CRISPR-Cas surveillance complex by anti-CRISPR protein AcrIF13.
J.Biol.Chem., 298, 2022
|
|
5WUK
 
 | |
7VIK
 
 | |
7XNO
 
 | |
7VLX
 
 | | Cryo-EM structures of Listeria monocytogenes man-PTS | | Descriptor: | Mannose/fructose/sorbose family PTS transporter subunit IIC, PTS mannose family transporter subunit IID, alpha-D-mannopyranose | | Authors: | Wang, J.W. | | Deposit date: | 2021-10-05 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Structural Basis of Pore Formation in the Mannose Phosphotransferase System by Pediocin PA-1.
Appl.Environ.Microbiol., 88, 2022
|
|
7VLY
 
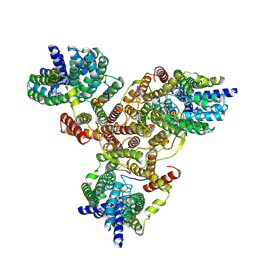 | | Cryo-EM structure of Listeria monocytogenes man-PTS complexed with pediocin PA-1 | | Descriptor: | Bacteriocin pediocin PA-1, Mannose/fructose/sorbose family PTS transporter subunit IIC, PTS mannose family transporter subunit IID, ... | | Authors: | Wang, J.W. | | Deposit date: | 2021-10-05 | | Release date: | 2021-12-01 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | Structural Basis of Pore Formation in the Mannose Phosphotransferase System by Pediocin PA-1.
Appl.Environ.Microbiol., 88, 2022
|
|
7VI9
 
 | |
7VII
 
 | |
7VIA
 
 | |
7FI4
 
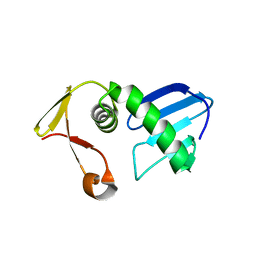 | | Structure of AcrIF13 | | Descriptor: | AcrIF13 | | Authors: | Feng, Y, Gao, T. | | Deposit date: | 2021-07-30 | | Release date: | 2022-07-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Mechanistic insights into the inhibition of the CRISPR-Cas surveillance complex by anti-CRISPR protein AcrIF13.
J.Biol.Chem., 298, 2022
|
|
7X27
 
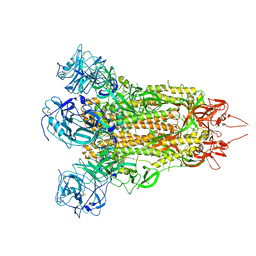 | | MERS-CoV spike complex | | Descriptor: | Spike glycoprotein | | Authors: | Zeng, J.W, Zhang, S.Y, Wang, X.W. | | Deposit date: | 2022-02-25 | | Release date: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | cryo-EM structures of a human neutralizing antibody bound to MERS-CoV spike glycoprotein
To Be Published
|
|
5WT1
 
 | |
5WT3
 
 | |
7WE6
 
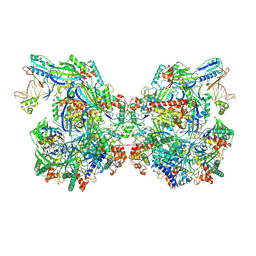 | | Structure of Csy-AcrIF24-dsDNA | | Descriptor: | AcrIF24, CRISPR type I-F/YPEST-associated protein Csy2, CRISPR-associated protein Csy3, ... | | Authors: | Zhang, L, Feng, Y. | | Deposit date: | 2021-12-22 | | Release date: | 2022-04-20 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Insights into the inhibition of type I-F CRISPR-Cas system by a multifunctional anti-CRISPR protein AcrIF24.
Nat Commun, 13, 2022
|
|
7ELM
 
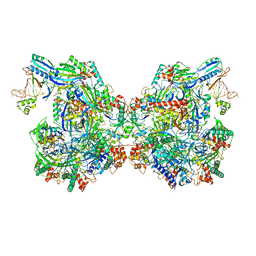 | | Structure of Csy-AcrIF24 | | Descriptor: | AcrIF24, CRISPR type I-F/YPEST-associated protein Csy2, CRISPR-associated protein Csy3, ... | | Authors: | Zhang, L, Feng, Y. | | Deposit date: | 2021-04-12 | | Release date: | 2022-04-20 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Insights into the inhibition of type I-F CRISPR-Cas system by a multifunctional anti-CRISPR protein AcrIF24.
Nat Commun, 13, 2022
|
|
7ELN
 
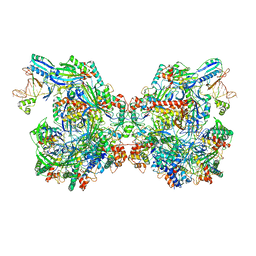 | | Structure of Csy-AcrIF24-dsDNA | | Descriptor: | 54-MER DNA, AcrIF24, CRISPR type I-F/YPEST-associated protein Csy2, ... | | Authors: | Zhang, L, Feng, Y. | | Deposit date: | 2021-04-12 | | Release date: | 2022-04-20 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Insights into the inhibition of type I-F CRISPR-Cas system by a multifunctional anti-CRISPR protein AcrIF24.
Nat Commun, 13, 2022
|
|
