2P2V
 
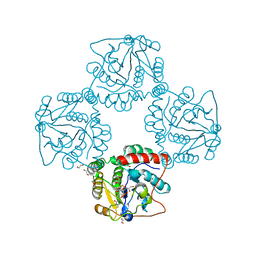 | | Crystal structure analysis of monofunctional alpha-2,3-sialyltransferase Cst-I from Campylobacter jejuni | | Descriptor: | 1,2-ETHANEDIOL, Alpha-2,3-sialyltransferase, CHLORIDE ION, ... | | Authors: | Chiu, C.P, Lairson, L.L, Gilbert, M, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C. | | Deposit date: | 2007-03-07 | | Release date: | 2007-07-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Analysis of the alpha-2,3-Sialyltransferase Cst-I from Campylobacter jejuni in Apo and Substrate-Analogue Bound Forms.
Biochemistry, 46, 2007
|
|
2CPU
 
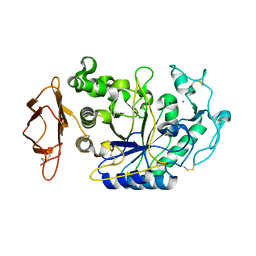 | | SUBSITE MAPPING OF THE ACTIVE SITE OF HUMAN PANCREATIC ALPHA-AMYLASE USING SUBSTRATES, THE PHARMACOLOGICAL INHIBITOR ACARBOSE, AND AN ACTIVE SITE VARIANT | | Descriptor: | ALPHA-AMYLASE, CALCIUM ION, CHLORIDE ION | | Authors: | Brayer, G.D, Sidhu, G, Maurus, R, Rydberg, E.H, Braun, C, Wang, Y, Nguyen, N.T, Overall, C.M, Withers, S.G. | | Deposit date: | 1999-06-08 | | Release date: | 2001-06-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Subsite mapping of the human pancreatic alpha-amylase active site through structural, kinetic, and mutagenesis techniques.
Biochemistry, 39, 2000
|
|
2F2H
 
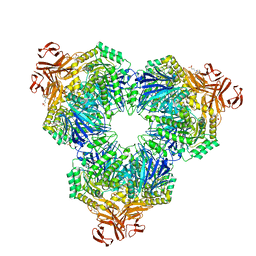 | | Structure of the YicI thiosugar Michaelis complex | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, 4-NITROPHENYL 6-THIO-6-S-ALPHA-D-XYLOPYRANOSYL-BETA-D-GLUCOPYRANOSIDE, GLYCEROL, ... | | Authors: | Kim, Y.-W, Lovering, A.L, Strynadka, N.C.J, Withers, S.G. | | Deposit date: | 2005-11-16 | | Release date: | 2006-02-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Expanding the Thioglycoligase Strategy to the Synthesis of alpha-linked Thioglycosides Allows Structural Investigation of the Parent Enzyme/Substrate Complex
J.Am.Chem.Soc., 128, 2006
|
|
2EXO
 
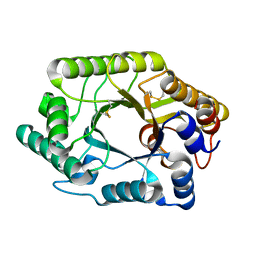 | | CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF THE BETA-1,4-GLYCANASE CEX FROM CELLULOMONAS FIMI | | Descriptor: | EXO-1,4-BETA-D-GLYCANASE | | Authors: | White, A, Withers, S.G, Gilkes, N.R, Rose, D.R. | | Deposit date: | 1994-07-11 | | Release date: | 1995-02-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the catalytic domain of the beta-1,4-glycanase cex from Cellulomonas fimi.
Biochemistry, 33, 1994
|
|
1FH8
 
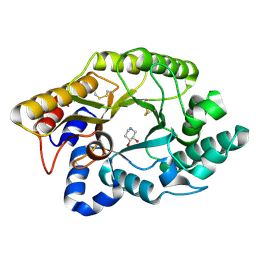 | | CRYSTAL STRUCTURE OF THE XYLANASE CEX WITH XYLOBIOSE-DERIVED ISOFAGOMINE INHIBITOR | | Descriptor: | BETA-1,4-XYLANASE, PIPERIDINE-3,4-DIOL, beta-D-xylopyranose | | Authors: | Notenboom, V, Williams, S.J, Hoos, R, Withers, S.G, Rose, D.R. | | Deposit date: | 2000-07-31 | | Release date: | 2000-08-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Detailed structural analysis of glycosidase/inhibitor interactions: complexes of Cex from Cellulomonas fimi with xylobiose-derived aza-sugars.
Biochemistry, 39, 2000
|
|
1FH7
 
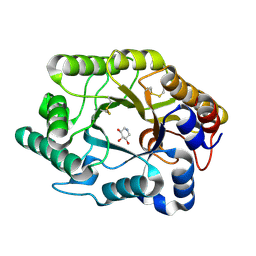 | | CRYSTAL STRUCTURE OF THE XYLANASE CEX WITH XYLOBIOSE-DERIVED INHIBITOR DEOXYNOJIRIMYCIN | | Descriptor: | BETA-1,4-XYLANASE, PIPERIDINE-3,4,5-TRIOL, beta-D-xylopyranose | | Authors: | Notenboom, V, Williams, S.J, Hoos, R, Withers, S.G, Rose, D.R. | | Deposit date: | 2000-07-31 | | Release date: | 2000-08-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Detailed structural analysis of glycosidase/inhibitor interactions: complexes of Cex from Cellulomonas fimi with xylobiose-derived aza-sugars.
Biochemistry, 39, 2000
|
|
1FHD
 
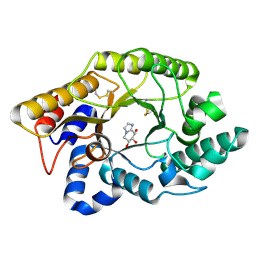 | | CRYSTAL STRUCTURE OF THE XYLANASE CEX WITH XYLOBIOSE-DERIVED IMIDAZOLE INHIBITOR | | Descriptor: | 5,6,7,8-TETRAHYDRO-IMIDAZO[1,2-A]PYRIDINE-6,7,8-TRIOL, BETA-1,4-XYLANASE, beta-D-xylopyranose | | Authors: | Notenboom, V, Williams, S.J, Hoos, R, Withers, S.G, Rose, D.R. | | Deposit date: | 2000-07-31 | | Release date: | 2000-08-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Detailed structural analysis of glycosidase/inhibitor interactions: complexes of Cex from Cellulomonas fimi with xylobiose-derived aza-sugars.
Biochemistry, 39, 2000
|
|
1FH9
 
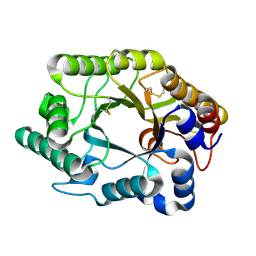 | | CRYSTAL STRUCTURE OF THE XYLANASE CEX WITH XYLOBIOSE-DERIVED LACTAM OXIME INHIBITOR | | Descriptor: | BETA-1,4-XYLANASE, beta-D-xylopyranose-(1-4)-(2Z,3S,4S,5R)-2-hydroxyiminopiperidine-3,4,5-triol | | Authors: | Notenboom, V, Williams, S.J, Hoos, R, Withers, S.G, Rose, D.R. | | Deposit date: | 2000-07-31 | | Release date: | 2000-08-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Detailed structural analysis of glycosidase/inhibitor interactions: complexes of Cex from Cellulomonas fimi with xylobiose-derived aza-sugars.
Biochemistry, 39, 2000
|
|
1G9R
 
 | | CRYSTAL STRUCTURE OF GALACTOSYLTRANSFERASE LGTC IN COMPLEX WITH MN AND UDP-2F-GALACTOSE | | Descriptor: | ACETIC ACID, GLYCOSYL TRANSFERASE, MANGANESE (II) ION, ... | | Authors: | Persson, K, Hoa, D.L, Diekelmann, M, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C.J. | | Deposit date: | 2000-11-27 | | Release date: | 2001-02-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the retaining galactosyltransferase LgtC from Neisseria meningitidis in complex with donor and acceptor sugar analogs.
Nat.Struct.Biol., 8, 2001
|
|
1GA8
 
 | | CRYSTAL STRUCTURE OF GALACOSYLTRANSFERASE LGTC IN COMPLEX WITH DONOR AND ACCEPTOR SUGAR ANALOGS. | | Descriptor: | 4-deoxy-beta-D-xylo-hexopyranose-(1-4)-beta-D-glucopyranose, GALACTOSYL TRANSFERASE LGTC, MANGANESE (II) ION, ... | | Authors: | Persson, K, Ly, H.D, Diekelmann, M, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C.J. | | Deposit date: | 2000-11-29 | | Release date: | 2001-02-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the retaining galactosyltransferase LgtC from Neisseria meningitidis in complex with donor and acceptor sugar analogs.
Nat.Struct.Biol., 8, 2001
|
|
1GVY
 
 | | Substrate distorsion by beta-mannanase from Pseudomonas cellulosa | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DINITROPHENYLENE, MANNAN ENDO-1,4-BETA-MANNOSIDASE, ... | | Authors: | Ducros, V, Zechel, D.L, Gilbert, H.J, Szabo, L, Withers, S.G, Davies, G.J. | | Deposit date: | 2002-02-28 | | Release date: | 2002-09-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Substrate Distortion by a Beta-Mannanase: Snapshots of the Michaelis and Covalent-Intermediate Complexes Suggest a B2,5 Conformation for the Transition State
Angew.Chem.Int.Ed.Engl., 41, 2002
|
|
1H6M
 
 | | Covalent glycosyl-enzyme intermediate of hen egg white lysozyme | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-deoxy-2-fluoro-alpha-D-glucopyranose, LYSOZYME C, SODIUM ION | | Authors: | Vocadlo, D.J, Davies, G.J, Laine, R, Withers, S.G. | | Deposit date: | 2001-06-19 | | Release date: | 2001-08-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Catalysis by Hen Egg-White Lysozyme Proceeds Via a Covalent Intermediate
Nature, 412, 2001
|
|
1GW1
 
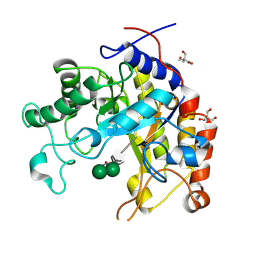 | | Substrate distortion by beta-mannanase from Pseudomonas cellulosa | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DINITROPHENYLENE, MANNAN ENDO-1,4-BETA-MANNOSIDASE, ... | | Authors: | Ducros, V, Zechel, D.L, Gilbert, H.J, Szabo, L, Withers, S.G, Davies, G.J. | | Deposit date: | 2002-03-01 | | Release date: | 2002-09-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Substrate Distortion by a Beta-Mannanase: Snapshots of the Michaelis and Covalent-Intermediate Complexes Suggest a B2,5 Conformation for the Transition State
Angew.Chem.Int.Ed.Engl., 41, 2002
|
|
1SS9
 
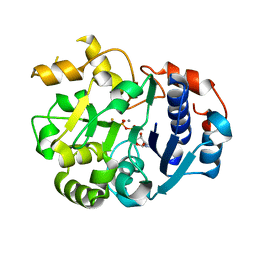 | | Crystal Structural Analysis of Active Site Mutant Q189E of LgtC | | Descriptor: | MANGANESE (II) ION, URIDINE-5'-DIPHOSPHATE-2-DEOXY-2-FLUOROGALACTOSE, alpha-1,4-galactosyl transferase | | Authors: | Lairson, L.L, Chiu, C.P, Ly, H.D, He, S, Wakarchuk, W.W, Strynadka, N.C, Withers, S.G. | | Deposit date: | 2004-03-23 | | Release date: | 2004-09-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Intermediate trapping on a mutant retaining alpha-galactosyltransferase identifies an unexpected aspartate residue.
J.Biol.Chem., 279, 2004
|
|
1S0I
 
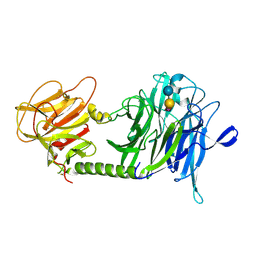 | | Trypanosoma cruzi trans-sialidase in complex with sialyl-lactose (Michaelis complex) | | Descriptor: | N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose, trans-sialidase | | Authors: | Amaya, M.F, Watts, A.G, Damager, I, Wehenkel, A, Nguyen, T, Buschiazzo, A, Paris, G, Frasch, A.C, Withers, S.G, Alzari, P.M. | | Deposit date: | 2003-12-31 | | Release date: | 2004-05-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Insights into the Catalytic Mechanism of Trypanosoma cruzi trans-Sialidase.
Structure, 12, 2004
|
|
1U33
 
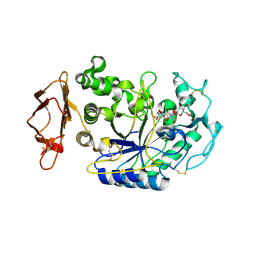 | | In situ extension as an approach for identifying novel alpha-amylase inhibitors | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4'-O-METHYL-MALTOSYL-ALPHA (1,4)-(Z, 3S,4S,5R,6R)-3,4,5-TRIHYDROXY-6-HYDROXYMETHYL-PIPERIDIN-2-ONE, ... | | Authors: | Numao, S, Li, C, Damager, I, Wrodnigg, T.M, Begum, A, Overall, C.M, Brayer, G.D, Withers, S.G. | | Deposit date: | 2004-07-20 | | Release date: | 2004-09-07 | | Last modified: | 2020-11-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | In Situ Extension as an Approach for Identifying Novel alpha-Amylase Inhibitors.
J.Biol.Chem., 279, 2004
|
|
1U30
 
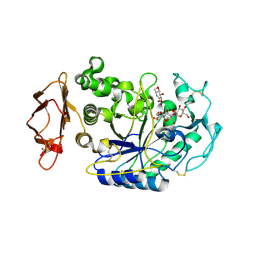 | | In situ extension as an approach for identifying novel alpha-amylase inhibitors, structure containing maltosyl-alpha (1,4)-D-gluconhydroximo-1,5-lactam | | Descriptor: | (2S,3S,4R,5R)-6-(HYDROXYAMINO)-2-(HYDROXYMETHYL)-2,3,4,5-TETRAHYDROPYRIDINE-3,4,5-TRIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-amylase, ... | | Authors: | Numao, S, Li, C, Damager, I, Wrodnigg, T.M, Begum, A, Overall, C.M, Brayer, G.D, Withers, S.G. | | Deposit date: | 2004-07-20 | | Release date: | 2004-09-07 | | Last modified: | 2020-11-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | In Situ Extension as an Approach for Identifying Novel alpha-Amylase Inhibitors.
J.Biol.Chem., 279, 2004
|
|
1RO8
 
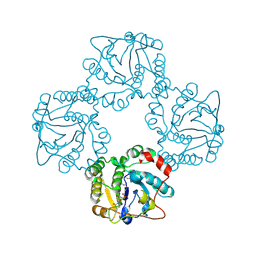 | | Structural analysis of the sialyltransferase CstII from Campylobacter jejuni in complex with a substrate analogue, cytidine-5'-monophosphate | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, alpha-2,3/8-sialyltransferase | | Authors: | Chiu, C.P, Watts, A.G, Lairson, L.L, Gilbert, M, Lim, D, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C. | | Deposit date: | 2003-12-01 | | Release date: | 2004-02-03 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural analysis of the sialyltransferase CstII from Campylobacter jejuni in complex with a substrate analog.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1S0J
 
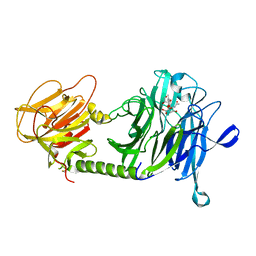 | | Trypanosoma cruzi trans-sialidase in complex with MuNANA (Michaelis complex) | | Descriptor: | 4-METHYL-2-OXO-2H-CHROMEN-7-YL 5-(ACETYLAMINO)-3,5-DIDEOXY-L-ERYTHRO-NON-2-ULOPYRANOSIDONIC ACID, trans-sialidase | | Authors: | Amaya, M.F, Watts, A.G, Damager, I, Wehenkel, A, Nguyen, T, Buschiazzo, A, Paris, G, Frasch, A.C, Withers, S.G, Alzari, P.M. | | Deposit date: | 2003-12-31 | | Release date: | 2004-05-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Insights into the Catalytic Mechanism of Trypanosoma cruzi trans-Sialidase.
Structure, 12, 2004
|
|
1UHV
 
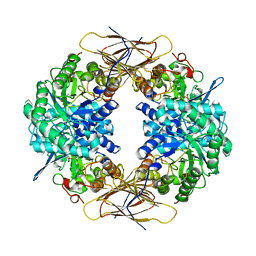 | | Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase | | Descriptor: | 1,5-anhydro-2-deoxy-2-fluoro-D-xylitol, Beta-xylosidase | | Authors: | Yang, J.K, Yoon, H.J, Ahn, H.J, Il Lee, B, Pedelacq, J.D, Liong, E.C, Berendzen, J, Laivenieks, M, Vieille, C, Zeikus, G.J, Vocadlo, D.J, Withers, S.G, Suh, S.W. | | Deposit date: | 2003-07-11 | | Release date: | 2003-12-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase.
J.Mol.Biol., 335, 2004
|
|
1UP7
 
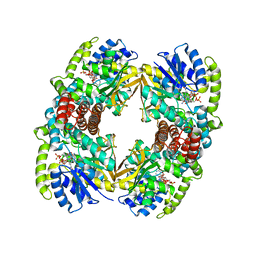 | | Structure of the 6-phospho-beta glucosidase from Thermotoga maritima at 2.4 Angstrom resolution in the tetragonal form with NAD and glucose-6-phosphate | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, 6-PHOSPHO-BETA-GLUCOSIDASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Varrot, A, Yip, V.L, Withers, S.G, Davies, G.J. | | Deposit date: | 2003-09-29 | | Release date: | 2004-11-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Nad+ and Metal-Ion Dependent Hydrolysis by Family 4 Glycosidases: Structural Insight Into Specificity for Phospho-Beta-D-Glucosides
J.Mol.Biol., 346, 2005
|
|
1V0M
 
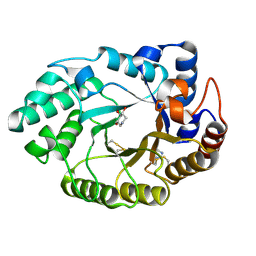 | | Xylanase Xyn10a from Streptomyces lividans in complex with xylobio-deoxynojirimycin at pH 7.5 | | Descriptor: | ENDO-1,4-BETA-XYLANASE A, IMIDAZOLE, PIPERIDINE-3,4,5-TRIOL, ... | | Authors: | Gloster, T.M, Williams, S.J, Roberts, S, Tarling, C.A, Wicki, J, Withers, S.G, Davies, G.J. | | Deposit date: | 2004-03-31 | | Release date: | 2004-08-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Atomic Resolution Analyses of the Binding of Xylobiose-Derived Deoxynojirimycin and Isofagomine to Xylanase Xyn10A
Chem.Commun.(Camb.), 16, 2004
|
|
1RO7
 
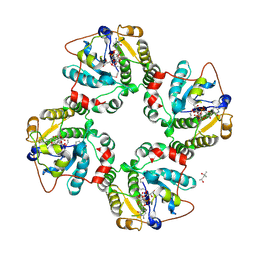 | | Structural analysis of the sialyltransferase CstII from Campylobacter jejuni in complex with a substrate analogue, CMP-3FNeuAc. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CYTIDINE-5'-MONOPHOSPHATE-3-FLUORO-N-ACETYL-NEURAMINIC ACID, alpha-2,3/8-sialyltransferase | | Authors: | Chiu, C.P, Watts, A.G, Lairson, L.L, Gilbert, M, Lim, D, Wakarchuk, W.W, Withers, S.G, Strynadka, N.C. | | Deposit date: | 2003-12-01 | | Release date: | 2004-02-03 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of the sialyltransferase CstII from Campylobacter jejuni in complex with a substrate analog.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1UP4
 
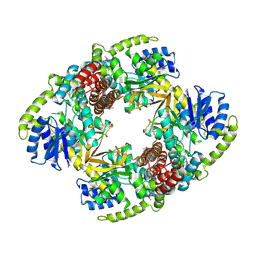 | |
1V0N
 
 | | Xylanase Xyn10a from Streptomyces lividans in complex with xylobio-isofagomine at pH 7.5 | | Descriptor: | 1,2-ETHANEDIOL, ENDO-1,4-BETA-XYLANASE A, IMIDAZOLE, ... | | Authors: | Gloster, T.M, Williams, S.J, Roberts, S, Tarling, C.A, Wicki, J, Withers, S.G, Davies, G.J. | | Deposit date: | 2004-03-31 | | Release date: | 2004-08-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic Resolution Analyses of the Binding of Xylobiose-Derived Deoxynojirimycin and Isofagomine to Xylanase Xyn10A
Chem.Commun.(Camb.), 16, 2004
|
|
