2YD4
 
 | | Crystal structure of the N-terminal Ig1-2 module of Chicken Receptor Protein Tyrosine Phosphatase Sigma | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, PROTEIN-TYROSINE PHOSPHATASE CRYPALPHA1 ISOFORM, ... | | Authors: | Coles, C.H, Shen, Y, Tenney, A.P, Siebold, C, Sutton, G.C, Lu, W, Gallagher, J.T, Jones, E.Y, Flanagan, J.G, Aricescu, A.R. | | Deposit date: | 2011-03-17 | | Release date: | 2011-04-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Proteoglycan-Specific Molecular Switch for Rptp Sigma Clustering and Neuronal Extension.
Science, 332, 2011
|
|
1O8T
 
 | | Global Structure and Dynamics of Human Apolipoprotein CII in Complex with Micelles: Evidence for increased mobility of the helix involved in the activation of lipoprotein lipase | | Descriptor: | APOLIPOPROTEIN C-II | | Authors: | Zdunek, J, Martinez, G.V, Schleucher, J, Lycksell, P.O, Yin, Y, Nilsson, S, Shen, Y, Olivecrona, G, Wijmenga, S. | | Deposit date: | 2002-11-29 | | Release date: | 2003-02-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Global Structure and Dynamics of Human Apolipoprotein Cii in Complex with Micelles: Evidence for Increased Mobility of the Helix Involved in the Activation of Lipoprotein Lipase
Biochemistry, 42, 2003
|
|
3O59
 
 | |
1XPV
 
 | | Solution Structure of Northeast Structural Genomics Target Protein XcR50 from X. Campestris | | Descriptor: | hypothetical protein XCC2852 | | Authors: | Shao, Y, Acton, T.B, Liu, G, Ma, L, Shen, Y, Xiao, R, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-10-09 | | Release date: | 2004-12-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Northeast Structural Genomics Target Protein XcR50 from X. Campestris
To be Published
|
|
1XN8
 
 | | Solution Structure of Bacillus subtilis Protein yqbG: The Northeast Structural Genomics Consortium Target SR215 | | Descriptor: | Hypothetical protein yqbG | | Authors: | Liu, G, Ma, L, Shen, Y, Acton, T, Atreya, H.S, Xiao, R, Joachimiak, A, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-10-04 | | Release date: | 2004-12-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR data collection and analysis protocol for high-throughput protein structure determination.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
3NK7
 
 | | Structure of the Nosiheptide-resistance methyltransferase S-adenosyl-L-methionine Complex | | Descriptor: | 23S rRNA methyltransferase, S-ADENOSYLMETHIONINE | | Authors: | Yang, H, Wang, Z, Shen, Y, Wang, P, Murchie, A, Xu, Y. | | Deposit date: | 2010-06-18 | | Release date: | 2010-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Nosiheptide-Resistance Methyltransferase of Streptomyces actuosus
Biochemistry, 49, 2010
|
|
3NK6
 
 | | Structure of the Nosiheptide-resistance methyltransferase | | Descriptor: | 23S rRNA methyltransferase | | Authors: | Yang, H, Wang, Z, Shen, Y, Wang, P, Murchie, A, Xu, Y. | | Deposit date: | 2010-06-18 | | Release date: | 2010-07-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Nosiheptide-Resistance Methyltransferase of Streptomyces actuosus
Biochemistry, 49, 2010
|
|
6U6P
 
 | |
6U6R
 
 | |
6U6S
 
 | |
1MI1
 
 | | Crystal Structure of the PH-BEACH Domain of Human Neurobeachin | | Descriptor: | Neurobeachin | | Authors: | Jogl, G, Shen, Y, Gebauer, D, Li, J, Wiegmann, K, Kashkar, H, Kroenke, M, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-08-21 | | Release date: | 2002-09-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the BEACH domain reveals an unusual fold and extensive association with a novel PH domain.
EMBO J., 21, 2002
|
|
4XFV
 
 | | Crystal Structure of Elp2 | | Descriptor: | Elongator complex protein 2 | | Authors: | Lin, Z, Dong, C, Long, J, Shen, Y. | | Deposit date: | 2014-12-29 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The elp2 subunit is essential for elongator complex assembly and functional regulation
Structure, 23, 2015
|
|
6U6Q
 
 | |
3EDT
 
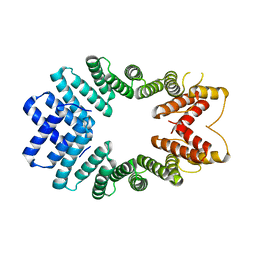 | | Crystal structure of the mutated S328N hKLC2 TPR domain | | Descriptor: | Kinesin light chain 2, UNKNOWN ATOM OR ION | | Authors: | Zhu, H, Shen, Y, MacKenzie, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-09-03 | | Release date: | 2009-02-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of mutated S328N TPR domain of human kinesin light chain 2
To be published
|
|
1OD2
 
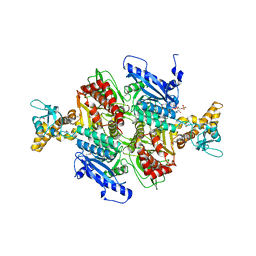 | | Acetyl-CoA Carboxylase Carboxyltransferase Domain | | Descriptor: | ACETYL COENZYME *A, ACETYL-COENZYME A CARBOXYLASE, ADENINE | | Authors: | Zhang, H, Yang, Z, Shen, Y, Tong, L. | | Deposit date: | 2003-02-12 | | Release date: | 2003-04-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the carboxyltransferase domain of acetyl-coenzyme A carboxylase.
Science, 299, 2003
|
|
1OD4
 
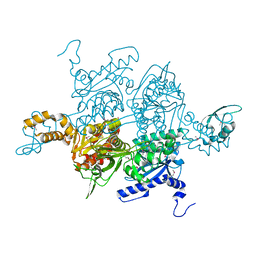 | | Acetyl-CoA Carboxylase Carboxyltransferase Domain | | Descriptor: | ACETYL-COENZYME A CARBOXYLASE, ADENINE | | Authors: | Zhang, H, Yang, Z, Shen, Y, Tong, L. | | Deposit date: | 2003-02-12 | | Release date: | 2003-04-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the carboxyltransferase domain of acetyl-coenzyme A carboxylase.
Science, 299, 2003
|
|
1S57
 
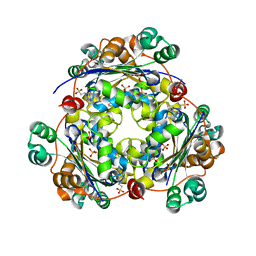 | | crystal structure of nucleoside diphosphate kinase 2 from Arabidopsis | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Nucleoside diphosphate kinase II, SULFATE ION | | Authors: | Im, Y.J, Kim, J.-I, Shen, Y, Na, Y, Han, Y.-J, Kim, S.-H, Song, P.-S, Eom, S.H. | | Deposit date: | 2004-01-20 | | Release date: | 2004-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of Arabidopsis thaliana nucleoside diphosphate kinase-2 for phytochrome-mediated light signaling
J.Mol.Biol., 343, 2004
|
|
2NZ0
 
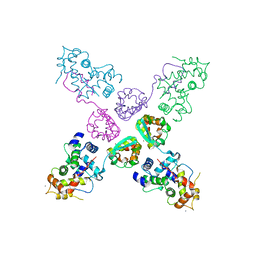 | | Crystal structure of potassium channel Kv4.3 in complex with its regulatory subunit KChIP1 | | Descriptor: | CALCIUM ION, Kv channel-interacting protein 1, Potassium voltage-gated channel subfamily D member 3, ... | | Authors: | Wang, H, Yan, Y, Shen, Y, Chen, L, Wang, K. | | Deposit date: | 2006-11-22 | | Release date: | 2006-12-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for modulation of Kv4 K(+) channels by auxiliary KChIP subunits.
Nat.Neurosci., 10, 2007
|
|
1S59
 
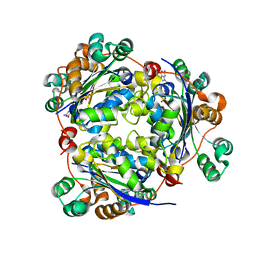 | | Structure of nucleoside diphosphate kinase 2 with bound dGTP from Arabidopsis | | Descriptor: | 2'-DEOXYGUANOSINE-5'-DIPHOSPHATE, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, Nucleoside diphosphate kinase II | | Authors: | Im, Y.J, Kim, J.-I, Shen, Y, Na, Y, Han, Y.-J, Kim, S.-H, Song, P.-S, Eom, S.H. | | Deposit date: | 2004-01-20 | | Release date: | 2004-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural analysis of Arabidopsis thaliana nucleoside diphosphate kinase-2 for phytochrome-mediated light signaling
J.Mol.Biol., 343, 2004
|
|
6VX9
 
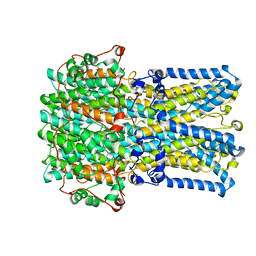 | | bestrophin-2 Ca2+- unbound state 1 (EGTA only) | | Descriptor: | Bestrophin, CHLORIDE ION | | Authors: | Owji, A.P, Zhao, Q, Ji, C, Kittredge, A, Hopiavuori, A, Fu, Z, Ward, N, Clarke, O, Shen, Y, Zhang, Y, Hendrickson, W.A, Yang, T. | | Deposit date: | 2020-02-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.17 Å) | | Cite: | Structural and functional characterization of the bestrophin-2 anion channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VX8
 
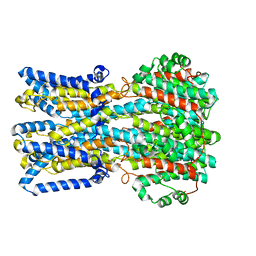 | | bestrophin-2 Ca2+- unbound state 2 (EGTA only) | | Descriptor: | Bestrophin, CHLORIDE ION | | Authors: | Owji, A.P, Zhao, Q, Ji, C, Kittredge, A, Hopiavuori, A, Fu, Z, Ward, N, Clarke, O, Shen, Y, Zhang, Y, Hendrickson, W.A, Yang, T. | | Deposit date: | 2020-02-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Structural and functional characterization of the bestrophin-2 anion channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VX5
 
 | | bestrophin-2 Ca2+- unbound state (250 nM Ca2+) | | Descriptor: | Bestrophin, CHLORIDE ION | | Authors: | Owji, A.P, Zhao, Q, Ji, C, Kittredge, A, Hopiavuori, A, Fu, Z, Ward, N, Clarke, O, Shen, Y, Zhang, Y, Hendrickson, W.A, Yang, T. | | Deposit date: | 2020-02-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structural and functional characterization of the bestrophin-2 anion channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VX6
 
 | | bestrophin-2 Ca2+-bound state (250 nM Ca2+) | | Descriptor: | Bestrophin, CALCIUM ION, CHLORIDE ION | | Authors: | Owji, A.P, Zhao, Q, Ji, C, Kittredge, A, Hopiavuori, A, Fu, Z, Ward, N, Clarke, O, Shen, Y, Zhang, Y, Hendrickson, W.A, Yang, T. | | Deposit date: | 2020-02-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and functional characterization of the bestrophin-2 anion channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VX7
 
 | | bestrophin-2 Ca2+-bound state (5 mM Ca2+) | | Descriptor: | Bestrophin, CALCIUM ION, CHLORIDE ION | | Authors: | Owji, A.P, Zhao, Q, Ji, C, Kittredge, A, Hopiavuori, A, Fu, Z, Ward, N, Clarke, O, Shen, Y, Zhang, Y, Hendrickson, W.A, Yang, T. | | Deposit date: | 2020-02-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.36 Å) | | Cite: | Structural and functional characterization of the bestrophin-2 anion channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
1ZNV
 
 | | How a His-metal finger endonuclease ColE7 binds and cleaves DNA with a transition metal ion cofactor | | Descriptor: | Colicin E7, Colicin E7 immunity protein, NICKEL (II) ION, ... | | Authors: | Doudeva, L.G, Huang, H, Hsia, K.C, Shi, Z, Li, C.L, Shen, Y, Yuan, H.S. | | Deposit date: | 2005-05-12 | | Release date: | 2006-03-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structural analysis and metal-dependent stability and activity studies of the ColE7 endonuclease domain in complex with DNA/Zn2+ or inhibitor/Ni2+
Protein Sci., 15, 2006
|
|
