8UL8
 
 | | LSD1-CoREST in complex with T15, short soaking | | Descriptor: | Lysine-specific histone demethylase 1A, REST corepressor 1, methyl 3-{(1R,3S,3aS,13R)-8-[(2S,3S,4R)-5-{[(S)-{[(S)-{[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}-2,3,4-trihydroxypentyl]-1-hydroxy-10,11-dimethyl-4,6-dioxo-2,3,4,5,6,8-hexahydro-1H-benzo[g]pyrrolo[2,1-e]pteridin-3-yl}benzoate (non-preferred name) | | Authors: | Caroli, J, Mattevi, A. | | Deposit date: | 2023-10-16 | | Release date: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Distal drug resistance mutations promote covalent inhibitor-adduct Grob fragmentation in LSD1
To Be Published
|
|
8UL6
 
 | | LSD1-CoREST in complex with T16, long soaking | | Descriptor: | Lysine-specific histone demethylase 1A, REST corepressor 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methyl (2R,3S,4S)-2,3,4-trihydroxy-5-[(1R,3S,3aS,13R)-1-hydroxy-10,11-dimethyl-3-[3-(methylcarbamoyl)phenyl]-4,6-dioxo-2,3,5,6-tetrahydro-1H-benzo[g]pyrrolo[2,1-e]pteridin-8(4H)-yl]pentyl dihydrogen diphosphate (non-preferred name) | | Authors: | Caroli, J, Mattevi, A. | | Deposit date: | 2023-10-16 | | Release date: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Distal drug resistance mutations promote covalent inhibitor-adduct Grob fragmentation in LSD1
To Be Published
|
|
8UNI
 
 | |
8UOM
 
 | |
8ULB
 
 | | LSD1-CoREST in complex with T17, long soaking | | Descriptor: | Lysine-specific histone demethylase 1A, REST corepressor 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methyl (2R,3S,4S)-5-[(1R,3S,3aS,13R)-3-[3-(dimethylcarbamoyl)phenyl]-1-hydroxy-10,11-dimethyl-4,6-dioxo-2,3,5,6-tetrahydro-1H-benzo[g]pyrrolo[2,1-e]pteridin-8(4H)-yl]-2,3,4-trihydroxypentyl dihydrogen diphosphate (non-preferred name) | | Authors: | Caroli, J, Mattevi, A. | | Deposit date: | 2023-10-16 | | Release date: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Distal drug resistance mutations promote covalent inhibitor-adduct Grob fragmentation in LSD1
To Be Published
|
|
8UMQ
 
 | | LSD1-CoREST in complex with T18, long soaking | | Descriptor: | Lysine-specific histone demethylase 1A, REST corepressor 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methyl (2R,3S,4S)-5-[(1R,3S,3aS,13R)-3-(3-benzamidophenyl)-1-hydroxy-10,11-dimethyl-4,6-dioxo-2,3,5,6-tetrahydro-1H-benzo[g]pyrrolo[2,1-e]pteridin-8(4H)-yl]-2,3,4-trihydroxypentyl dihydrogen diphosphate (non-preferred name) | | Authors: | Caroli, J, Mattevi, A. | | Deposit date: | 2023-10-18 | | Release date: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (3.26 Å) | | Cite: | Distal drug resistance mutations promote covalent inhibitor-adduct Grob fragmentation in LSD1
To Be Published
|
|
8CAP
 
 | |
8CAL
 
 | |
8CAO
 
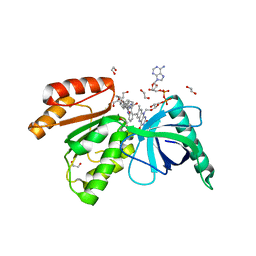 | | Crystal structure of dehydrogenase domain of Cylindrospermum stagnale NADPH-Oxidase 5 (NOX5) in complex with CA24 | | Descriptor: | 1,2-ETHANEDIOL, 8-[2-(4-cyclohexylphenyl)quinolin-4-yl]carbonyl-1,3,8-triazaspiro[4.5]decane-2,4-dione, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Reis, J, Mattevi, A. | | Deposit date: | 2023-01-24 | | Release date: | 2023-09-27 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Targeting ROS production through inhibition of NADPH oxidases.
Nat.Chem.Biol., 19, 2023
|
|
8CB0
 
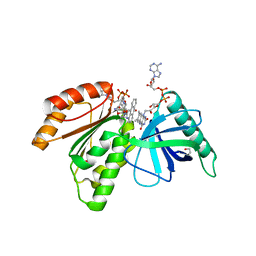 | | Crystal structure of dehydrogenase domain of Cylindrospermum stagnale NADPH-Oxidase 5 (NOX5) in complex with M41 and NADP+ | | Descriptor: | 1,2-ETHANEDIOL, 15-(1,4-dioxa-8-azaspiro[4.5]decan-8-yl)-14-azatetracyclo[7.7.1.0^{2,7}.0^{13,17}]heptadeca-1(16),2(7),3,5,9,11,13(17),14-octaen-8-one, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Reis, J, Mattevi, A. | | Deposit date: | 2023-01-24 | | Release date: | 2023-09-27 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Targeting ROS production through inhibition of NADPH oxidases.
Nat.Chem.Biol., 19, 2023
|
|
8CAK
 
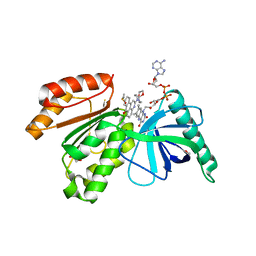 | | Crystal structure of dehydrogenase domain of Cylindrospermum stagnale NADPH-Oxidase 5 (NOX5) in complex with M41 | | Descriptor: | 1,2-ETHANEDIOL, 15-(1,4-dioxa-8-azaspiro[4.5]decan-8-yl)-14-azatetracyclo[7.7.1.0^{2,7}.0^{13,17}]heptadeca-1(16),2(7),3,5,9,11,13(17),14-octaen-8-one, DIMETHYL SULFOXIDE, ... | | Authors: | Reis, J, Mattevi, A. | | Deposit date: | 2023-01-24 | | Release date: | 2023-09-27 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Targeting ROS production through inhibition of NADPH oxidases.
Nat.Chem.Biol., 19, 2023
|
|
7AL4
 
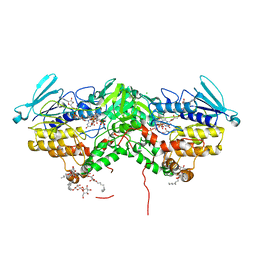 | | Ancestral Flavin-containing monooxygenase (FMO) 1 (mammalian) | | Descriptor: | Ancestral Flavin-containing monooxygenase 1 (mammalian), CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Nicoll, C.R, Mattevi, A. | | Deposit date: | 2020-10-05 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Ancestral reconstruction of mammalian FMO1 enables structural determination, revealing unique features that explain its catalytic properties.
J.Biol.Chem., 296, 2020
|
|
2VFS
 
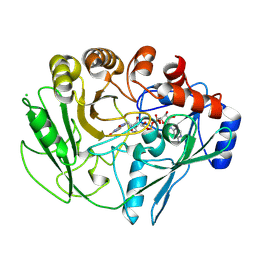 | |
2VFU
 
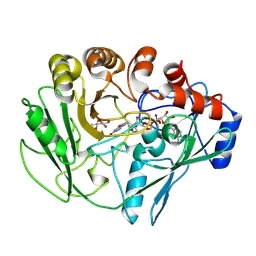 | |
2V6J
 
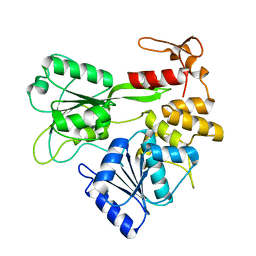 | | Kokobera Virus Helicase: Mutant Met47Thr | | Descriptor: | RNA HELICASE | | Authors: | Speroni, S, De Colibus, L, Coutard, B, Canard, B, Mattevi, A. | | Deposit date: | 2007-07-18 | | Release date: | 2008-03-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Biochemical Analysis of Kokobera Virus Helicase
Proteins: Struct., Funct., Bioinf., 70, 2008
|
|
7O1R
 
 | |
7O1X
 
 | |
7O2G
 
 | |
7O1Z
 
 | |
7O2D
 
 | |
6HQG
 
 | | Cytochrome P450-153 from Phenylobacterium zucineum | | Descriptor: | Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Fiorentini, F, Mattevi, A. | | Deposit date: | 2018-09-25 | | Release date: | 2018-12-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Extreme Structural Plasticity in the CYP153 Subfamily of P450s Directs Development of Designer Hydroxylases.
Biochemistry, 57, 2018
|
|
6HQD
 
 | | Cytochrome P450-153 from Pseudomonas sp. 19-rlim | | Descriptor: | Cytochrome P450, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Fiorentini, F, Mattevi, A. | | Deposit date: | 2018-09-24 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Extreme Structural Plasticity in the CYP153 Subfamily of P450s Directs Development of Designer Hydroxylases.
Biochemistry, 57, 2018
|
|
6HQW
 
 | |
2VGG
 
 | | HUMAN ERYTHROCYTE PYRUVATE KINASE: R479H MUTANT | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 2-PHOSPHOGLYCOLIC ACID, MANGANESE (II) ION, ... | | Authors: | Valentini, G, Chiarelli, L.R, Fortin, R, Dolzan, M, Galizzi, A, Abraham, D.J, Wang, C, Bianchi, P, Zanella, A, Mattevi, A. | | Deposit date: | 2007-11-13 | | Release date: | 2007-11-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structure and Function of Human Erythrocyte Pyruvate Kinase. Molecular Basis of Nonspherocytic Hemolytic Anemia.
J.Biol.Chem., 277, 2002
|
|
2VGF
 
 | | HUMAN ERYTHROCYTE PYRUVATE KINASE: T384M mutant | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 2-PHOSPHOGLYCOLIC ACID, MANGANESE (II) ION, ... | | Authors: | Valentini, G, Chiarelli, L.R, Fortin, R, Dolzan, M, Galizzi, A, Abraham, D.J, Wang, C, Bianchi, P, Zanella, A, Mattevi, A. | | Deposit date: | 2007-11-12 | | Release date: | 2007-11-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure and Function of Human Erythrocyte Pyruvate Kinase. Molecular Basis of Nonspherocytic Hemolytic Anemia.
J.Biol.Chem., 277, 2002
|
|
