4ARO
 
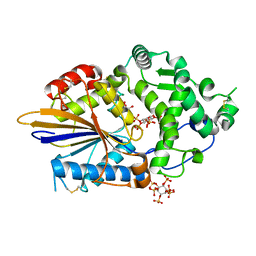 | | Hafnia Alvei phytase in complex with myo-inositol hexakis sulphate | | Descriptor: | D-MYO-INOSITOL-HEXASULPHATE, DI(HYDROXYETHYL)ETHER, HISTIDINE ACID PHOSPHATASE, ... | | Authors: | Moroz, O.V, Blagova, E.B, Ariza, A, Turkenburg, J.P, Vevodova, J, Roberts, S, Vind, J, Sjoholm, C, Lassen, S.F, De Maria, L, Glitsoe, V, Skov, L.K, Wilson, K.S. | | Deposit date: | 2012-04-25 | | Release date: | 2013-05-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Degradation of Phytate by the 6-Phytase from Hafnia Alvei: A Combined Structural and Solution Study.
Plos One, 8, 2013
|
|
1OYU
 
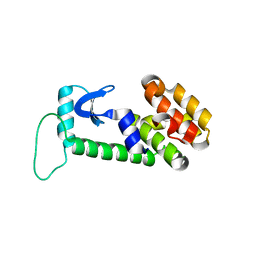 | |
8V32
 
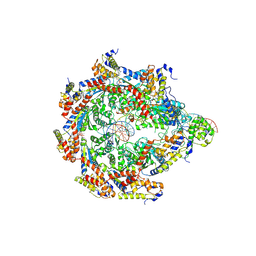 | | TnsD-TnsC-DNA complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (41-MER), MAGNESIUM ION, ... | | Authors: | Chang, L, Wang, S. | | Deposit date: | 2023-11-26 | | Release date: | 2024-10-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structure of TnsABCD transpososome reveals mechanisms of targeted DNA transposition.
Cell, 2024
|
|
8B32
 
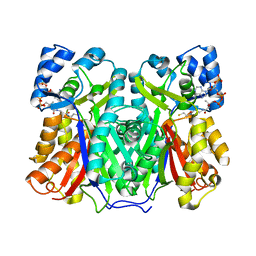 | |
1DZ5
 
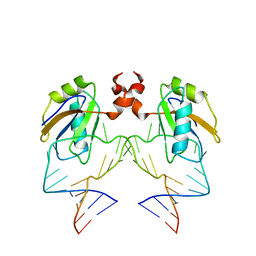 | | The NMR structure of the 38KDa U1A protein-PIE RNA complex reveals the basis of cooperativity in regulation of polyadenylation by human U1A protein | | Descriptor: | PIE, RNA (5'-R(*GP*AP*GP*AP*CP*AP*UP*UP*GP*CP*AP*CP*CP* CP*GP*GP*AP*GP*UP*CP*UP*C)-3'), U1 SMALL NUCLEAR RIBONUCLEOPROTEIN A | | Authors: | Varani, L, Gunderson, S.I, Mattaj, I.W, Kay, L.E, Neuhaus, D, Varani, G. | | Deposit date: | 2000-02-16 | | Release date: | 2000-03-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The NMR Structure of the 38kDa U1A Protein-Pie RNA Complex Reveals the Basis of Cooperativity in Regulation of Polyadenylation by Human U1A Protein
Nat.Struct.Biol., 7, 2000
|
|
8U9L
 
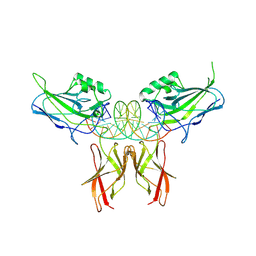 | | Crystal Structure of RelA-cRel chimera complex with DNA | | Descriptor: | DNA (5'-D(P*GP*AP*AP*TP*CP*GP*GP*AP*AP*AP*TP*TP*CP*CP*CP*AP*TP*CP*AP*A)-3'), DNA (5'-D(P*TP*TP*GP*AP*TP*GP*GP*GP*AP*AP*TP*TP*TP*CP*CP*GP*AP*TP*TP*C)-3'), Transcription factor p65,Proto-oncogene c-Rel chimera | | Authors: | Chang, A, Wu, Y, Li, S.X, Smale, S, Chen, L. | | Deposit date: | 2023-09-19 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (3.091 Å) | | Cite: | Crystal Structure of RelA-cRel chimera complex with DNA
To Be Published
|
|
8B35
 
 | |
5NOB
 
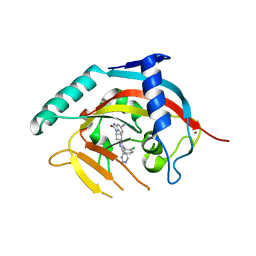 | | Crystal structure of human tankyrase 2 in complex with OD336 | | Descriptor: | 1-[3-[4-(2-chlorophenyl)-5-pyrimidin-4-yl-1,2,4-triazol-3-yl]cyclobutyl]-2-oxidanylidene-3~{H}-benzimidazole-5-carbonitrile, Tankyrase-2, ZINC ION | | Authors: | Ignatev, A, Lehtio, L. | | Deposit date: | 2017-04-11 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of a Novel Series of Tankyrase Inhibitors by a Hybridization Approach.
J. Med. Chem., 60, 2017
|
|
4B3U
 
 | | Pseudomonas aeruginosa RmlA in complex with allosteric inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-amino-1-benzyl-5-(ethylamino)pyrimidine-2,4(1H,3H)-dione, CHLORIDE ION, ... | | Authors: | Alphey, M.S, Pirrie, L, Torrie, L.S, Gardiner, M, Sarkar, A, Brenk, R, Westwood, N.J, Gray, D, Naismith, J.H. | | Deposit date: | 2012-07-26 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Allosteric competitive inhibitors of the glucose-1-phosphate thymidylyltransferase (RmlA) from Pseudomonas aeruginosa.
ACS Chem. Biol., 8, 2013
|
|
8B3C
 
 | | Chalcone synthase from Hordeum vulgare complexed with CoA and eriodictyol | | Descriptor: | (2S)-2-(3,4-DIHYDROXYPHENYL)-5,7-DIHYDROXY-2,3-DIHYDRO-4H-CHROMEN-4-ONE, COENZYME A, Chalcone synthase 2 | | Authors: | Zhang, L, Groves, M.R. | | Deposit date: | 2022-09-16 | | Release date: | 2023-09-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Engineering a Plant Polyketide Synthase for the Biosynthesis of Methylated Flavonoids.
J.Agric.Food Chem., 72, 2024
|
|
4AAW
 
 | | S.pneumoniae GlmU in complex with an antibacterial inhibitor | | Descriptor: | 4-{[1-(2-{[({5-[(3-carboxypropanoyl)amino]-2,4-dimethoxyphenyl}sulfonyl)amino]methyl}phenyl)piperidin-4-yl]methoxy}-4-oxobutanoic acid, BIFUNCTIONAL PROTEIN GLMU, SULFATE ION | | Authors: | Otterbein, L, Breed, J, Ogg, D.J. | | Deposit date: | 2011-12-05 | | Release date: | 2012-02-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inhibitors of Acetyltransferase Domain of N-Acetylglucosamine-1-Phosphate-Uridyltransferase/ Glucosamine-1-Phosphate-Acetyltransferase (Glmu). Part 1: Hit to Lead Evaluation of a Novel Arylsulfonamide Series.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
8UF8
 
 | |
5N77
 
 | | Crystal structure of the cytosolic domain of the CorA magnesium channel from Escherichia coli in complex with magnesium | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, MAGNESIUM ION, Magnesium transport protein CorA | | Authors: | Lerche, M, Sandhu, H, Flockner, L, Hogbom, M, Rapp, M. | | Deposit date: | 2017-02-20 | | Release date: | 2017-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and Cooperativity of the Cytosolic Domain of the CorA Mg(2+) Channel from Escherichia coli.
Structure, 25, 2017
|
|
1DQE
 
 | | BOMBYX MORI PHEROMONE BINDING PROTEIN | | Descriptor: | HEXADECA-10,12-DIEN-1-OL, PHEROMONE-BINDING PROTEIN | | Authors: | Sandler, B.H, Nikonova, L, Leal, W.S, Clardy, J. | | Deposit date: | 2000-01-04 | | Release date: | 2001-01-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Sexual attraction in the silkworm moth: structure of the pheromone-binding-protein-bombykol complex.
Chem.Biol., 7, 2000
|
|
3W0I
 
 | | Crystal Structure of Rat VDR Ligand Binding Domain in Complex with Novel Nonsecosteroidal Ligands | | Descriptor: | (2S)-3-{4-[3-(4-{[(2R)-2-hydroxy-3,3-dimethylbutyl]oxy}phenyl)pentan-3-yl]phenoxy}propane-1,2-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 Receptor | | Authors: | Shimizu, T, Asano, L, Kuwabara, N, Ito, I, Waku, T, Yanagisawa, J, Miyachi, H. | | Deposit date: | 2012-10-30 | | Release date: | 2013-10-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for vitamin D receptor agonism by novel non-secosteroidal ligands.
Febs Lett., 587, 2013
|
|
6FW4
 
 | | Protein-protein interactions and conformational changes : Importance of the hydrophobic cavity of TolA C-terminal domain | | Descriptor: | TolA protein | | Authors: | Navarro, R, van Heijenoort, C, Bornet, O, Houot, L, Lloubes, R, Guerlesquin, F, Nouailler, M. | | Deposit date: | 2018-03-05 | | Release date: | 2019-03-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Induced fit conformational changes of Vibrio cholerae TolAIII domain during the complex formation with the viral PIIIN1 domain: Structural and High-pressure NMR studies.
To Be Published
|
|
8RUW
 
 | |
4A29
 
 | | Structure of the engineered retro-aldolase RA95.0 | | Descriptor: | 1-(6-METHOXYNAPHTHALEN-2-YL)BUTANE-1,3-DIONE, D-MALATE, ENGINEERED RETRO-ALDOL ENZYME RA95.0 | | Authors: | Giger, L, Caner, S, Kast, P, Baker, D, Ban, N, Hilvert, D. | | Deposit date: | 2011-09-23 | | Release date: | 2012-11-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Evolution of a designed retro-aldolase leads to complete active site remodeling.
Nat.Chem.Biol., 9, 2013
|
|
3V7V
 
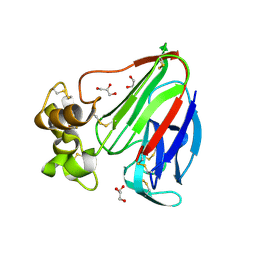 | | Thaumatin by Classical Hanging Drop Vapour Diffusion after 1.81 MGy X-Ray dose at ESRF ID29 beamline (Best Case) | | Descriptor: | GLYCEROL, Thaumatin I | | Authors: | Belmonte, L, Scudieri, D, Tripathi, S, Pechkova, E, Nicolini, C. | | Deposit date: | 2011-12-22 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Langmuir-Blodgett nanotemplate and radiation resistance in protein crystals: state of the art.
CRIT.REV.EUKARYOT.GENE EXPR., 22, 2012
|
|
8B5W
 
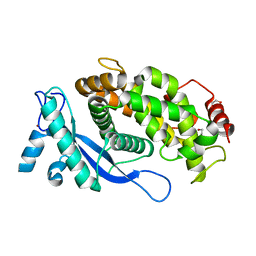 | | Crystal structure of the E3 module from UBR4 | | Descriptor: | 1,2-ETHANEDIOL, ZINC ION, cDNA FLJ12511 fis, ... | | Authors: | Virdee, S, Mabbitt, P.D, Barnsby-Greer, L. | | Deposit date: | 2022-09-24 | | Release date: | 2023-10-04 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | UBE2A and UBE2B are recruited by an atypical E3 ligase module in UBR4.
Nat.Struct.Mol.Biol., 31, 2024
|
|
4A2R
 
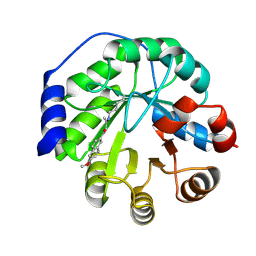 | | Structure of the engineered retro-aldolase RA95.5-5 | | Descriptor: | 1-(6-METHOXYNAPHTHALEN-2-YL)BUTANE-1,3-DIONE, INDOLE-3-GLYCEROL PHOSPHATE SYNTHASE | | Authors: | Giger, L, Caner, S, Kast, P, Baker, D, Ban, N, Hilvert, D. | | Deposit date: | 2011-09-28 | | Release date: | 2012-11-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.302 Å) | | Cite: | Evolution of a designed retro-aldolase leads to complete active site remodeling.
Nat.Chem.Biol., 9, 2013
|
|
4A3R
 
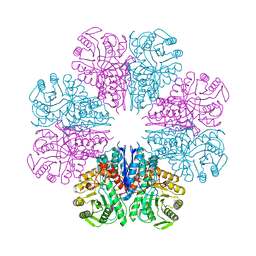 | | Crystal structure of Enolase from Bacillus subtilis. | | Descriptor: | CITRIC ACID, ENOLASE, SODIUM ION | | Authors: | Newman, J.A, Hewitt, L, Rodrigues, C, Solovyova, A.S, Harwood, C.R, Lewis, R.J. | | Deposit date: | 2011-10-04 | | Release date: | 2012-08-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dissection of the Network of Interactions that Links RNA Processing with Glycolysis in the Bacillus Subtilis Degradosome.
J.Mol.Biol., 416, 2012
|
|
4A3S
 
 | | Crystal structure of PFK from Bacillus subtilis | | Descriptor: | 6-PHOSPHOFRUCTOKINASE | | Authors: | Newman, J.A, Hewitt, L, Rodrigues, C, Solovyova, A.S, Harwood, C.R, Lewis, R.J. | | Deposit date: | 2011-10-04 | | Release date: | 2012-08-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Dissection of the Network of Interactions that Links RNA Processing with Glycolysis in the Bacillus Subtilis Degradosome.
J.Mol.Biol., 416, 2012
|
|
8B6M
 
 | |
5YL9
 
 | |
