5MQT
 
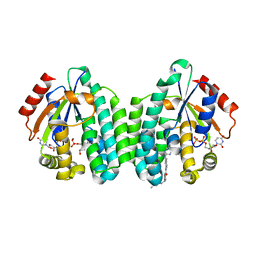 | | Crystal structure of dCK mutant C3S in complex with imatinib and UDP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, Deoxycytidine kinase, ... | | Authors: | Saez-Ayala, M, Rebuffet, E, Hammam, K, Gros, L, Lopez, S, Hajem, B, Humbert, M, Baudelet, E, Audebert, S, Betzi, S, Lugari, A, Combes, S, Pez, D, Letard, S, Mansfield, C, Moussy, A, de Sepulveda, P, Morelli, X, Dubreuil, P. | | Deposit date: | 2016-12-20 | | Release date: | 2017-11-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Dual protein kinase and nucleoside kinase modulators for rationally designed polypharmacology.
Nat Commun, 8, 2017
|
|
4C40
 
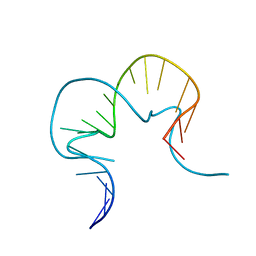 | |
5MR5
 
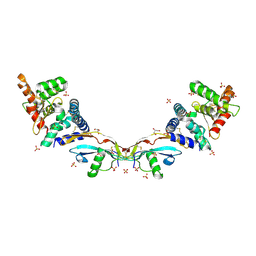 | | Ligand-receptor complex. | | Descriptor: | GDNF family receptor alpha-2, GLYCEROL, Neurturin, ... | | Authors: | Sandmark, J, Oster, L, Aagaard, A, Roth, R, Dahl, G. | | Deposit date: | 2016-12-21 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and biophysical characterization of the human full-length neurturin-GFRa2 complex: A role for heparan sulfate in signaling.
J. Biol. Chem., 293, 2018
|
|
5YR3
 
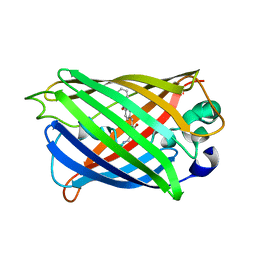 | | Structure of sfYFP66BPA | | Descriptor: | Yellow fluorescent protein | | Authors: | Kang, F, Wang, L, Wang, J. | | Deposit date: | 2017-11-08 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | sfYFP66BPA
To Be Published
|
|
4C45
 
 | | Crystal structure of human pterin-4-alpha-carbinolamine dehydratase 2 (PCBD2) | | Descriptor: | 1,2-ETHANEDIOL, PTERIN-4-ALPHA-CARBINOLAMINE DEHYDRATASE 2 | | Authors: | Kopec, J, Kiyani, W, Vollmar, M, Shrestha, L, Canning, P, von Delft, F, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Yue, W.W. | | Deposit date: | 2013-08-30 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structure of Human Pterin-4-Alpha- Carbinolamine Dehydratase 2 (Pcbd2)
To be Published
|
|
5XYY
 
 | | The structure of p38 alpha in complex with a triazol inhibitor | | Descriptor: | 3-(5-{[(2-chloro-6-fluorophenyl)methyl]amino}-4H-1,2,4-triazol-3-yl)phenol, Mitogen-activated protein kinase 14 | | Authors: | Wang, Y.L, Sun, Y.Z, Cao, R, Liu, D, Li, L, Qi, X.B, Huang, N. | | Deposit date: | 2017-07-11 | | Release date: | 2018-01-17 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | In Silico Identification of a Novel Hinge-Binding Scaffold for Kinase Inhibitor Discovery.
J. Med. Chem., 60, 2017
|
|
6F47
 
 | | Crystal structure of the catalytic domain of botulinum neurotoxin X | | Descriptor: | Catalytic domain of botulinum neurotoxin X, ZINC ION | | Authors: | Masuyer, G, Henriksson, L, Kosenina, S, Zhang, S, Barkho, S, Shen, Y, Dong, M, Stenmark, P. | | Deposit date: | 2017-11-29 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural characterisation of the catalytic domain of botulinum neurotoxin X - high activity and unique substrate specificity.
Sci Rep, 8, 2018
|
|
5MK4
 
 | | Crystal structure of the Retinoid X Receptor alpha in complex with synthetic honokiol derivative 7 and a fragment of the TIF2 co-activator. | | Descriptor: | (~{E})-3-[3-(2-methyl-5-phenyl-phenyl)-4-oxidanyl-phenyl]prop-2-enoic acid, CHLORIDE ION, Nuclear receptor coactivator 2, ... | | Authors: | Andrei, S.A, Scheepstra, M, Brunsveld, L, Ottmann, C. | | Deposit date: | 2016-12-02 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ligand Dependent Switch from RXR Homo- to RXR-NURR1 Heterodimerization.
ACS Chem Neurosci, 8, 2017
|
|
3T95
 
 | | Crystal structure of LsrB from Yersinia pestis complexed with autoinducer-2 | | Descriptor: | (2R,4S)-2-methyl-2,3,3,4-tetrahydroxytetrahydrofuran, Autoinducer 2-binding protein lsrB | | Authors: | Kavanaugh, J.S, Gakhar, L, Horswill, A.R. | | Deposit date: | 2011-08-02 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The structure of LsrB from Yersinia pestis complexed with autoinducer-2.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
4C06
 
 | | Crystal structure of M. musculus protein arginine methyltransferase PRMT6 with MgCl2 | | Descriptor: | MAGNESIUM ION, PROTEIN ARGININE N-METHYLTRANSFERASE 6 | | Authors: | Bonnefond, L, Cura, V, Troffer-Charlier, N, Mailliot, J, Wurtz, J.M, Cavarelli, J. | | Deposit date: | 2013-07-31 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Functional Insights from High Resolution Structures of Mouse Protein Arginine Methyltransferase 6.
J.Struct.Biol., 191, 2015
|
|
1F4N
 
 | | C2 CRYSTAL STRUCTURE OF ALA2ILE2-6, A VERSION OF ROP WITH A REPACKED HYDROPHOBIC CORE AND A NEW FOLD. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ROP ALA2ILE2-6 | | Authors: | Willis, M.A, Bishop, B, Regan, L, Brunger, A.T. | | Deposit date: | 2000-06-08 | | Release date: | 2001-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Dramatic structural and thermodynamic consequences of repacking a protein's hydrophobic core.
Structure Fold.Des., 8, 2000
|
|
1OEX
 
 | | Atomic Resolution Structure of Endothiapepsin in Complex with a Hydroxyethylene Transition State Analogue Inhibitor H261 | | Descriptor: | ENDOTHIAPEPSIN, GLYCEROL, INHIBITOR H261, ... | | Authors: | Coates, L, Erskine, P.T, Mall, S, Gill, R.S, Wood, S.P, Myles, D.A.A, Cooper, J.B. | | Deposit date: | 2003-03-31 | | Release date: | 2003-04-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic Resolution Analysis of the Catalytic Site of an Aspartic Proteinase and an Unexpected Mode of Binding by Short Peptides
Protein Sci., 12, 2003
|
|
8B64
 
 | | Cryo-EM structure of RC-LH1-PufX photosynthetic core complex from Rba. capsulatus | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, ... | | Authors: | Bracun, L, Yamagata, A, Shirouzu, M, Liu, L.N. | | Deposit date: | 2022-09-26 | | Release date: | 2023-02-01 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.589 Å) | | Cite: | Cryo-EM structure of a monomeric RC-LH1-PufX supercomplex with high-carotenoid content from Rhodobacter capsulatus.
Structure, 31, 2023
|
|
1EZK
 
 | | Crystal structure of recombinant tryparedoxin I | | Descriptor: | TRYPAREDOXIN I | | Authors: | Hofmann, B, Guerrero, S.A, Kalisz, H.M, Menge, U, Nogoceke, E, Montemartini, M, Singh, M, Flohe, L, Hecht, H.J. | | Deposit date: | 2000-05-11 | | Release date: | 2000-05-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of tryparedoxins revealing interaction with trypanothione.
Biol.Chem., 382, 2001
|
|
6EWK
 
 | | T. californica AChE in complex with a 3-hydroxy-2-pyridine aldoxime. | | Descriptor: | 2-[(~{E})-hydroxyiminomethyl]-6-(5-morpholin-4-ylpentyl)pyridin-3-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, ... | | Authors: | de la Mora, E, Weik, M, Braiki, A, Mougeot, R, Jean, L, Renard, P.I. | | Deposit date: | 2017-11-04 | | Release date: | 2018-11-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Potent 3-Hydroxy-2-Pyridine Aldoxime Reactivators of Organophosphate-Inhibited Cholinesterases with Predicted Blood-Brain Barrier Penetration.
Chemistry, 24, 2018
|
|
1OEW
 
 | | ATOMIC RESOLUTION STRUCTURE OF NATIVE ENDOTHIAPEPSIN | | Descriptor: | ENDOTHIAPEPSIN, GLYCEROL, SERINE, ... | | Authors: | Coates, L, Erskine, P.T, Mall, S, Gill, R.S, Wood, S.P, Myles, D.A.A, Cooper, J.B. | | Deposit date: | 2003-03-31 | | Release date: | 2003-04-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Atomic Resolution Analysis of the Catalytic Site of an Aspartic Proteinase and an Unexpected Mode of Binding by Short Peptides
Protein Sci., 12, 2003
|
|
4CAY
 
 | | Crystal structure of a human Anp32e-H2A.Z-H2B complex | | Descriptor: | ACIDIC LEUCINE-RICH NUCLEAR PHOSPHOPROTEIN 32 FAMILY MEMBER E, HISTONE H2A.Z, HISTONE H2B TYPE 1-J | | Authors: | Obri, A, Ouararhni, K, Papin, C, Diebold, M.-L, Padmanabhan, K, Marek, M, Stoll, I, Roy, L, Reilly, P.T, Mak, T.W, Dimitrov, S, Romier, C, Hamiche, A. | | Deposit date: | 2013-10-09 | | Release date: | 2014-01-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Anp32E is a Histone Chaperone that Removes H2A.Z from Chromatin
Nature, 648, 2014
|
|
1F1X
 
 | | CRYSTAL STRUCTURE OF HOMOPROTOCATECHUATE 2,3-DIOXYGENASE FROM BREVIBACTERIUM FUSCUM | | Descriptor: | HOMOPROTOCATECHUATE 2,3-DIOXYGENASE, HYDRATED FE | | Authors: | Vetting, M.W, Lipscomb, J.D, Wackett, L.P, Que Jr, L, Ohlendorf, D.H. | | Deposit date: | 2000-05-20 | | Release date: | 2003-06-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic comparison of manganese- and iron-dependent homoprotocatechuate 2,3-dioxygenases.
J.Bacteriol., 186, 2004
|
|
4BUD
 
 | | Crystal structure of human tankyrase 2 in complex with 2-(4-tert- butylphenyl)-1,4-dihydroquinazolin-4-one | | Descriptor: | 2-(4-tert-butylphenyl)-1,4-dihydroquinazolin-4-one, GLYCEROL, SULFATE ION, ... | | Authors: | Haikarainen, T, Narwal, M, Lehtio, L. | | Deposit date: | 2013-06-20 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Para-Substituted 2-Phenyl-3,4-Dihydroquinazolin-4-Ones as Potent and Selective Tankyrase Inhibitors.
Chemmedchem, 8, 2013
|
|
3ST9
 
 | | Crystal structure of ClpP in heptameric form from Staphylococcus aureus | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, CALCIUM ION, GLYCEROL, ... | | Authors: | Zhang, J, Ye, F, Lan, L, Jiang, H, Luo, C, Yang, C.-G. | | Deposit date: | 2011-07-09 | | Release date: | 2011-09-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural switching of Staphylococcus aureus Clp protease: a key to understanding protease dynamics
J.Biol.Chem., 286, 2011
|
|
1F08
 
 | | CRYSTAL STRUCTURE OF THE DNA-BINDING DOMAIN OF THE REPLICATION INITIATION PROTEIN E1 FROM PAPILLOMAVIRUS | | Descriptor: | BROMIDE ION, REPLICATION PROTEIN E1 | | Authors: | Enemark, E.J, Chen, G, Vaughn, D.E, Stenlund, A, Joshua-Tor, L. | | Deposit date: | 2000-05-15 | | Release date: | 2001-05-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the DNA binding domain of the replication initiation protein E1 from papillomavirus.
Mol.Cell, 6, 2000
|
|
5MPW
 
 | | Crystal structure of Arabidopsis thaliana RNA editing factor MORF1 | | Descriptor: | BROMIDE ION, Multiple organellar RNA editing factor 1, mitochondrial, ... | | Authors: | Haag, S, Schindler, M, Berndt, L, Brennicke, A, Takenaka, M, Weber, G. | | Deposit date: | 2016-12-19 | | Release date: | 2017-02-22 | | Last modified: | 2017-12-27 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Crystal structures of the Arabidopsis thaliana organellar RNA editing factors MORF1 and MORF9.
Nucleic Acids Res., 45, 2017
|
|
3SWM
 
 | | The NAC domain of ANAC019 in complex with DNA, gold derivative | | Descriptor: | GOLD ION, NAC domain-containing protein 19, oligonucleotide forward, ... | | Authors: | Welner, D, Lo Leggio, L. | | Deposit date: | 2011-07-14 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (4.25 Å) | | Cite: | DNA binding by the plant-specific NAC transcription factors in crystal and solution: a firm link to WRKY and GCM transcription factors.
Biochem.J., 444, 2012
|
|
6F1Z
 
 | | Roadblock-1 region of the dynein tail/dynactin/BICDR1 complex | | Descriptor: | Cytoplasmic dynein 1 intermediate chain 2, Dynein light chain roadblock-type 1 | | Authors: | Urnavicius, L, Lau, C.K, Elshenawy, M.M, Morales-Rios, E, Motz, C, Yildiz, A, Carter, A.P. | | Deposit date: | 2017-11-23 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM shows how dynactin recruits two dyneins for faster movement.
Nature, 554, 2018
|
|
1OXC
 
 | | LecB (PA-LII) in complex with FUCOSE | | Descriptor: | CALCIUM ION, SULFATE ION, alpha-L-fucopyranose, ... | | Authors: | Loris, R, Tielker, D, Jaeger, K.-E, Wyns, L. | | Deposit date: | 2003-04-02 | | Release date: | 2003-09-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural Basis of Carbohydrate Recognition by the Lectin LecB from Pseudomonas aeruginosa
J.MOL.BIOL., 331, 2003
|
|
