6TI7
 
 | | Mixing Abeta(1-40) and Abeta(1-42) peptides generates unique amyloid fibrils | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Cerofolini, L, Ravera, E, Bologna, S, Wiglenda, T, Boddrich, A, Purfurst, B, Benilova, A, Korsak, M, Gallo, G, Rizzo, D, Gonnelli, L, Fragai, M, De Strooper, B, Wanker, E.E, Luchinat, C. | | Deposit date: | 2019-11-21 | | Release date: | 2020-07-22 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | Mixing A beta (1-40) and A beta (1-42) peptides generates unique amyloid fibrils.
Chem.Commun.(Camb.), 56, 2020
|
|
6TOS
 
 | | Crystal structure of the Orexin-1 receptor in complex with GSK1059865 | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, CITRIC ACID, Orexin receptor type 1, ... | | Authors: | Rappas, M, Ali, A, Bennett, K.A, Brown, J.D, Bucknell, S.J, Congreve, M, Cooke, R.M, Cseke, G, de Graaf, C, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Mason, J.S, Mould, R, Patel, J.C, Tehan, B.G, Weir, M, Christopher, J.A. | | Deposit date: | 2019-12-11 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Comparison of Orexin 1 and Orexin 2 Ligand Binding Modes Using X-ray Crystallography and Computational Analysis.
J.Med.Chem., 63, 2020
|
|
6TQ6
 
 | | Crystal structure of the Orexin-1 receptor in complex with Compound 14 | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, 2-(5-methylsulfonylpyridin-3-yl)-1,1-bis(oxidanylidene)-4-[[2,4,6-tris(fluoranyl)phenyl]methyl]pyrido[2,3-e][1,2,4]thiadiazin-3-one, Orexin receptor type 1, ... | | Authors: | Rappas, M, Ali, A, Bennett, K.A, Brown, J.D, Bucknell, S.J, Congreve, M, Cooke, R.M, Cseke, G, de Graaf, C, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Mason, J.S, Mould, R, Patel, J.C, Tehan, B.G, Weir, M, Christopher, J.A. | | Deposit date: | 2019-12-16 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.546 Å) | | Cite: | Comparison of Orexin 1 and Orexin 2 Ligand Binding Modes Using X-ray Crystallography and Computational Analysis.
J.Med.Chem., 63, 2020
|
|
6TUK
 
 | | Crystal structure of Fdr9 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, Putative oxidoreductase, ... | | Authors: | Rodriguez, A, Kluenemann, T, Blankenfeldt, W, Schallmey, A. | | Deposit date: | 2020-01-07 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Expression, purification and crystal structure determination of a ferredoxin reductase from the actinobacterium Thermobifida fusca.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
1JYQ
 
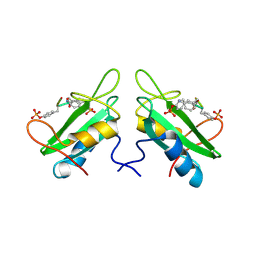 | | Xray Structure of Grb2 SH2 Domain Complexed with a Highly Affine Phospho Peptide | | Descriptor: | GROWTH FACTOR RECEPTOR-BOUND PROTEIN 2, mAZ-pY-(alpha Me)pY-N-NH2 peptide inhibitor | | Authors: | Nioche, P, Liu, W.-Q, Broutin, I, Charbonnier, F, Latreille, M.-T, Vidal, M, Roques, B, Garbay, C, Ducruix, A. | | Deposit date: | 2001-09-13 | | Release date: | 2002-03-13 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the SH2 domain of Grb2: highlight on the binding of a new high-affinity inhibitor.
J.Mol.Biol., 315, 2002
|
|
3E2C
 
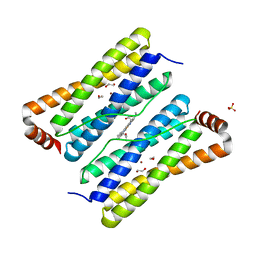 | | Escherichia coli Bacterioferritin Mutant E128R/E135R | | Descriptor: | 1,2-ETHANEDIOL, Bacterioferritin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Wong, S.G, Tom-Yew, S.A.L, Lewin, A, Le Brun, N.E, Moore, G.R, Murphy, M.E.P, Mauk, A.G. | | Deposit date: | 2008-08-05 | | Release date: | 2009-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Mechanistic Studies of a Stabilized Subunit Dimer Variant of Escherichia coli Bacterioferritin Identify Residues Required for Core Formation.
J.Biol.Chem., 284, 2009
|
|
3E47
 
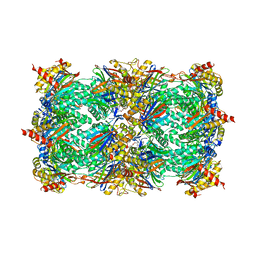 | | Crystal Structure of the Yeast 20S Proteasome in Complex with Homobelactosin C | | Descriptor: | Proteasome component C1, Proteasome component C11, Proteasome component C5, ... | | Authors: | Groll, M, Larionov, O.V, de Meijere, A. | | Deposit date: | 2008-08-10 | | Release date: | 2008-09-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Inhibitor-binding mode of homobelactosin C to proteasomes: new insights into class I MHC ligand generation
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
3E89
 
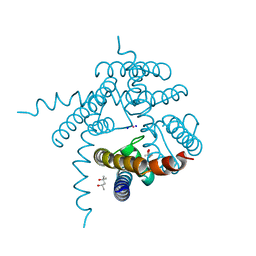 | |
3E8G
 
 | |
3EUW
 
 | |
3EAP
 
 | | Crystal structure of the RhoGAP domain of ARHGAP11A | | Descriptor: | Rho GTPase-activating protein 11A, UNKNOWN ATOM OR ION | | Authors: | Shen, Y, Shen, L, Tong, Y, Tempel, W, MacKenzie, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-08-26 | | Release date: | 2008-09-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the RhoGAP domain of ARHGAP11A
To be Published
|
|
1JTP
 
 | | Degenerate interfaces in antigen-antibody complexes | | Descriptor: | FORMIC ACID, LYSOZYME C, SODIUM ION, ... | | Authors: | Decanniere, K, Transue, T.R, Desmyter, A, Maes, D, Muyldermans, S, Wyns, L. | | Deposit date: | 2001-08-21 | | Release date: | 2001-12-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Degenerate interfaces in antigen-antibody complexes.
J.Mol.Biol., 313, 2001
|
|
1JAH
 
 | | H-RAS P21 PROTEIN MUTANT G12P, COMPLEXED WITH GUANOSINE-5'-[BETA,GAMMA-METHYLENE] TRIPHOSPHATE AND MAGNESIUM | | Descriptor: | C-HA-RAS, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER | | Authors: | Schweins, T, Scheffzek, K, Assheuer, R, Wittinghofer, A. | | Deposit date: | 1996-12-15 | | Release date: | 1997-07-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The role of the metal ion in the p21ras catalysed GTP-hydrolysis: Mn2+ versus Mg2+.
J.Mol.Biol., 266, 1997
|
|
1JAI
 
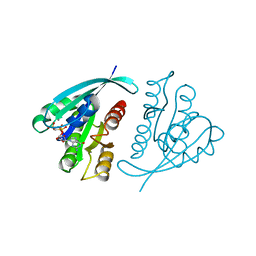 | | H-RAS P21 PROTEIN MUTANT G12P, COMPLEXED WITH GUANOSINE-5'-[BETA,GAMMA-METHYLENE] TRIPHOSPHATE AND MANGANESE | | Descriptor: | C-HA-RAS, MANGANESE (II) ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER | | Authors: | Schweins, T, Scheffzek, K, Assheuer, R, Wittinghofer, A. | | Deposit date: | 1996-12-15 | | Release date: | 1997-07-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The role of the metal ion in the p21ras catalysed GTP-hydrolysis: Mn2+ versus Mg2+.
J.Mol.Biol., 266, 1997
|
|
3E8E
 
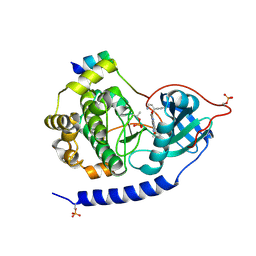 | | Crystal structures of the kinase domain of PKA in complex with ATP-competitive inhibitors | | Descriptor: | 4-[2-(4-amino-2,5-dihydro-1,2,5-oxadiazol-3-yl)-6-{[(1S)-3-amino-1-phenylpropyl]oxy}-1-ethyl-1H-imidazo[4,5-c]pyridin-4-yl]-2-methylbut-3-yn-2-ol, PKI inhibitor peptide, cAMP-dependent protein kinase catalytic subunit alpha | | Authors: | Concha, N.O, Elkins, P.A, Smallwood, A, Ward, P. | | Deposit date: | 2008-08-19 | | Release date: | 2008-11-18 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Aminofurazans as potent inhibitors of AKT kinase
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3EOS
 
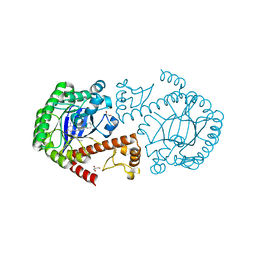 | | tRNA-guanine transglycosylase in complex with 6-amino-4-{2-[(cyclohexylmethyl)amino]ethyl}-2-(methylamino)-1,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one | | Descriptor: | 6-amino-4-{2-[(cyclohexylmethyl)amino]ethyl}-2-(methylamino)-1,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Ritschel, T, Heine, A, Klebe, G. | | Deposit date: | 2008-09-29 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | How to Replace the Residual Solvation Shell of Polar Active Site Residues to Achieve Nanomolar Inhibition of tRNA-Guanine Transglycosylase
Chemmedchem, 4, 2009
|
|
3EE6
 
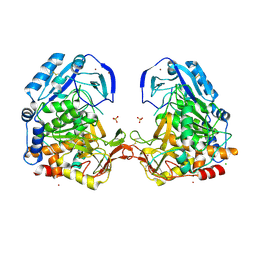 | | Crystal Structure Analysis of Tripeptidyl peptidase -I | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Pal, A, Kraetzner, R, Grapp, M, Gruene, T, Schreiber, K, Granborg, M, Urlaub, H, Asif, A.R, Becker, S, Gartner, J, Sheldrick, G.M, Steinfeld, R. | | Deposit date: | 2008-09-04 | | Release date: | 2008-11-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of tripeptidyl-peptidase I provides insight into the molecular basis of late infantile neuronal ceroid lipofuscinosis
J.Biol.Chem., 284, 2009
|
|
3NAD
 
 | | Crystal Structure of Phenolic Acid Decarboxylase from Bacillus pumilus UI-670 | | Descriptor: | Ferulate decarboxylase, SULFATE ION | | Authors: | Matte, A, Grosse, S, Bergeron, H, Abokitse, K, Lau, P.C.K. | | Deposit date: | 2010-06-01 | | Release date: | 2010-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural analysis of Bacillus pumilus phenolic acid decarboxylase, a lipocalin-fold enzyme.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
1K6X
 
 | | Crystal structure of Nmra, a negative transcriptional regulator in complex with NAD at 1.5 A resolution (Trigonal form) | | Descriptor: | CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, NmrA | | Authors: | Stammers, D.K, Ren, J, Leslie, K, Nichols, C.E, Lamb, H.K, Cocklin, S, Dodds, A, Hawkins, A.R. | | Deposit date: | 2001-10-17 | | Release date: | 2002-02-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structure of the negative transcriptional regulator NmrA reveals a structural superfamily which includes the short-chain dehydrogenase/reductases.
EMBO J., 20, 2002
|
|
4CK4
 
 | | Ovine beta-Lactoglobulin at Atomic Resolution | | Descriptor: | ACETATE ION, AMMONIUM ION, BETA_LACTOGLOBULIN-1/B, ... | | Authors: | Kontopidis, G, Nordle, A, Sawyer, L. | | Deposit date: | 2013-12-27 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Ovine Beta-Lactoglobulin at Atomic Resolution
Acta Crystallogr.,Sect.F, 70, 2014
|
|
4CQH
 
 | | Structure of Infrared Fluorescent Protein IFP2.0 | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, BACTERIOPHYTOCHROME, SODIUM ION | | Authors: | Lafaye, C, Yu, D, Noirclerc-Savoye, M, Shu, X, Royant, A. | | Deposit date: | 2014-02-17 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | An Improved Monomeric Infrared Fluorescent Protein for Neuronal and Tumour Brain Imaging.
Nat.Commun., 5, 2014
|
|
3EIE
 
 | | Crystal Structure of S.cerevisiae Vps4 in the SO4-bound state | | Descriptor: | SULFATE ION, Vacuolar protein sorting-associated protein 4 | | Authors: | Gonciarz, M.D, Whitby, F.G, Eckert, D.M, Kieffer, C, Heroux, A, Sundquist, W.I, Hill, C.P. | | Deposit date: | 2008-09-15 | | Release date: | 2008-09-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Biochemical and structural studies of yeast vps4 oligomerization.
J.Mol.Biol., 384, 2008
|
|
3EIP
 
 | | CRYSTAL STRUCTURE OF COLICIN E3 IMMUNITY PROTEIN: AN INHIBITOR TO A RIBOSOME-INACTIVATING RNASE | | Descriptor: | PROTEIN (COLICIN E3 IMMUNITY PROTEIN), ZINC ION | | Authors: | Li, C, Zhao, D, Djebli, A, Shoham, M. | | Deposit date: | 1999-03-29 | | Release date: | 1999-11-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of colicin E3 immunity protein: an inhibitor of a ribosome-inactivating RNase.
Structure Fold.Des., 7, 1999
|
|
3ED9
 
 | | Carbonmonoxy Sperm Whale Myoglobin at 140 K: Laser on [30 min] | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Tomita, A, Sato, T, Ichiyanagi, K, Nozawa, S, Ichikawa, H, Chollet, M, Kawai, F, Park, S.-Y, Koshihara, S, Adachi, S. | | Deposit date: | 2008-09-02 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Visualizing breathing motion of internal cavities in concert with ligand migration in myoglobin.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4CRW
 
 | | Complex of human DDX6 (RECA-C) and CNOT1 (MIF4G) | | Descriptor: | CCR4-NOT TRANSCRIPTION COMPLEX SUBUNIT 1, GLYCEROL, PROBABLE ATP-DEPENDENT RNA HELICASE DDX6 | | Authors: | Chen, Y, Boland, A, Izaurralde, E, Weichenrieder, O. | | Deposit date: | 2014-03-01 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Ddx6-Cnot1 Complex and W-Binding Pockets in Cnot9 Reveal Direct Links between Mirna Target Recognition and Silencing
Mol.Cell, 54, 2014
|
|
