4B4M
 
 | | Pseudomonas aeruginosa RmlA in complex with allosteric inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, ... | | Authors: | Alphey, M.S, Pirrie, L, Torrie, L.S, Gardiner, M, Westwood, N.J, Gray, D, Naismith, J.H. | | Deposit date: | 2012-07-31 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Allosteric competitive inhibitors of the glucose-1-phosphate thymidylyltransferase (RmlA) from Pseudomonas aeruginosa.
ACS Chem. Biol., 8, 2013
|
|
5M0T
 
 | | Alpha-ketoglutarate-dependent non-heme iron oxygenase EasH | | Descriptor: | 2-OXOGLUTARIC ACID, Alpha-ketoglutarate-dependent non-heme iron oxygenase EasH, FE (II) ION, ... | | Authors: | Jakubczyk, D, Caputi, L, Stevenson, C.E.M, Lawson, D.M, O'Connor, S.E. | | Deposit date: | 2016-10-05 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural characterization of EasH (Aspergillus japonicus) - an oxidase involved in cycloclavine biosynthesis.
Chem. Commun. (Camb.), 52, 2016
|
|
3TGK
 
 | | TRYPSINOGEN MUTANT D194N AND DELETION OF ILE 16-VAL 17 COMPLEXED WITH BOVINE PANCREATIC TRYPSIN INHIBITOR (BPTI) | | Descriptor: | CALCIUM ION, PANCREATIC TRYPSIN INHIBITOR, SULFATE ION, ... | | Authors: | Pasternak, A, White, A, Jeffery, C.J, Medina, N, Cahoon, M, Ringe, D, Hedstrom, L. | | Deposit date: | 1998-07-19 | | Release date: | 2001-07-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The energetic cost of induced fit catalysis: Crystal structures of trypsinogen mutants with enhanced activity and inhibitor affinity.
Protein Sci., 10, 2001
|
|
3TGV
 
 | | Crystal structure of HutZ,the heme storsge protein from Vibrio cholerae | | Descriptor: | BENZOIC ACID, Heme-binding protein HutZ | | Authors: | Liu, X, Gong, J, Wang, Z, Du, Q, Wei, T, Zhu, D, Huang, Y, Xu, S, Gu, L. | | Deposit date: | 2011-08-17 | | Release date: | 2012-08-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Crystal structure of HutZ,the heme storsge protein from Vibrio cholerae
To be Published
|
|
8AZU
 
 | | Paired helical tau filaments from high-spin supernatants of aqueous extracts from Alzheimer's disease brains | PHF Tau | | Descriptor: | Microtubule-associated protein tau | | Authors: | Yang, Y, Stern, M.A, Meunier, L.A, Liu, W, Cai, Y.Q, Ericsson, M, Liu, L, Selkoe, J.D, Goedert, M, Scheres, H.W.S. | | Deposit date: | 2022-09-06 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Abundant A beta fibrils in ultracentrifugal supernatants of aqueous extracts from Alzheimer's disease brains.
Neuron, 111, 2023
|
|
8Q3M
 
 | | Structure of Nucleosome Core with a Bound Kaposi Sarcoma Associated Herpesvirus LANA Peptide Having a Methionine to Ornithine Substitution | | Descriptor: | DNA (145-MER), Histone H2A type 1-B/E, Histone H2B type 1-K, ... | | Authors: | De Falco, L, Batchelor, L.K, Dyson, P.J, Davey, C.A. | | Deposit date: | 2023-08-04 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Viral peptide conjugates for metal-warhead delivery to chromatin.
Rsc Adv, 14, 2024
|
|
4BD0
 
 | | X-ray structure of a perdeuterated Toho-1 R274N R276N double mutant Beta-lactamase in complex with a fully deuterated boronic acid (BZB) | | Descriptor: | BENZO[B]THIOPHENE-2-BORONIC ACID, BETA-LACTAMASE TOHO-1, SULFATE ION | | Authors: | Tomanicek, S.J, Weiss, K.L, Standaert, R.F, Ostermann, A, Schrader, T.E, Ng, J.D, Coates, L. | | Deposit date: | 2012-10-04 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.207 Å) | | Cite: | Neutron and X-Ray Crystal Structures of a Perdeuterated Enzyme Inhibitor Complex Reveal the Catalytic Proton Network of the Toho-1 Beta-Lactamase for the Acylation Reaction.
J.Biol.Chem., 288, 2013
|
|
1ORW
 
 | | Crystal Structure of Porcine Dipeptidyl Peptidase IV (CD26) in Complex with a Peptidomimetic Inhibitor | | Descriptor: | (2S)-PYRROLIDIN-2-YLMETHYLAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Engel, M, Hoffmann, T, Wagner, L, Wermann, M, Heiser, U, Kiefersauer, R, Huber, R, Bode, W, Demuth, H.U, Brandstetter, H. | | Deposit date: | 2003-03-16 | | Release date: | 2003-05-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | The Crystal Structure of Dipeptidyl Peptidase IV (CD26) Reveals its Functional Regulation and Enzymatic Mechanism
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
8Q36
 
 | | Structure of Nucleosome Core with a Bound Metallopeptide Conjugate (Foamy Virus GAG Peptide-Au[I] Compound) | | Descriptor: | DNA (145-MER), GAG structural protein, Histone H2A type 1-B/E, ... | | Authors: | De Falco, L, Batchelor, L.K, Dyson, P.J, Davey, C.A. | | Deposit date: | 2023-08-03 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Viral peptide conjugates for metal-warhead delivery to chromatin.
Rsc Adv, 14, 2024
|
|
8TNL
 
 | |
1P8V
 
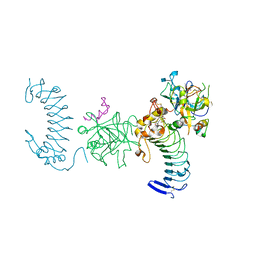 | | CRYSTAL STRUCTURE OF THE COMPLEX OF PLATELET RECEPTOR GPIB-ALPHA AND ALPHA-THROMBIN AT 2.6A | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIISOPROPYL PHOSPHONATE, ... | | Authors: | Dumas, J.J, Kumar, R, Seehra, J, Somers, W.S, Mosyak, L. | | Deposit date: | 2003-05-07 | | Release date: | 2003-07-22 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the GpIbalpha-Thrombin Complex Essential for Platelet Aggregation
Science, 301, 2003
|
|
1CJA
 
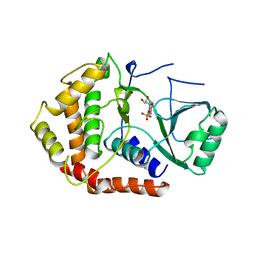 | | ACTIN-FRAGMIN KINASE, CATALYTIC DOMAIN FROM PHYSARUM POLYCEPHALUM | | Descriptor: | ADENOSINE MONOPHOSPHATE, PROTEIN (ACTIN-FRAGMIN KINASE) | | Authors: | Steinbacher, S, Hof, P, Eichinger, L, Schleicher, M, Gettemans, J, Vandekerckhove, J, Huber, R, Benz, J. | | Deposit date: | 1999-04-08 | | Release date: | 1999-06-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of the Physarum polycephalum actin-fragmin kinase: an atypical protein kinase with a specialized substrate-binding domain.
EMBO J., 18, 1999
|
|
6EYD
 
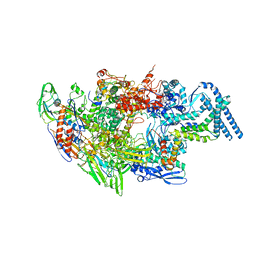 | | Structure of Mycobacterium smegmatis RNA polymerase Sigma-A holoenzyme | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Kouba, T, Barvik, I, Krasny, L. | | Deposit date: | 2017-11-11 | | Release date: | 2018-12-12 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.22 Å) | | Cite: | The Core and Holoenzyme Forms of RNA Polymerase fromMycobacterium smegmatis.
J. Bacteriol., 201, 2019
|
|
8Q3X
 
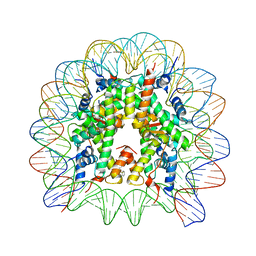 | | Structure of Nucleosome Core with a Bound Metallopeptide Conjugate (Kaposi Sarcoma Associated Herpesvirus LANA Peptide-Au[I] Compound) | | Descriptor: | 4-diphenylphosphanylbenzoic acid, DNA (145-MER), GOLD ION, ... | | Authors: | De Falco, L, Batchelor, L.K, Dyson, P.J, Davey, C.A. | | Deposit date: | 2023-08-04 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Viral peptide conjugates for metal-warhead delivery to chromatin.
Rsc Adv, 14, 2024
|
|
1P9K
 
 | | THE SOLUTION STRUCTURE OF YBCJ FROM E. COLI REVEALS A RECENTLY DISCOVERED ALFAL MOTIF INVOLVED IN RNA-BINDING | | Descriptor: | orf, hypothetical protein | | Authors: | Volpon, L, Lievre, C, Osborne, M.J, Gandhi, S, Iannuzzi, P, Larocque, R, Matte, A, Cygler, M, Gehring, K, Ekiel, I, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2003-05-12 | | Release date: | 2003-11-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The solution structure of YbcJ from Escherichia coli reveals a recently discovered alphaL motif involved in RNA binding.
J.Bacteriol., 185, 2003
|
|
1OXE
 
 | | Expansion of the Genetic Code Enables Design of a Novel "Gold" Class of Green Fluorescent Proteins | | Descriptor: | cyan fluorescent protein cfp | | Authors: | Hyun Bae, J, Rubini, M, Jung, G, Wiegand, G, Seifert, M.H, Azim, M.K, Kim, J.S, Zumbusch, A, Holak, T.A, Moroder, L, Huber, R, Budisa, N. | | Deposit date: | 2003-04-02 | | Release date: | 2003-12-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Expansion of the Genetic Code Enables Design of a Novel "Gold" Class of Green Fluorescent Proteins
J.Mol.Biol., 328, 2003
|
|
8TOA
 
 | | CryoEM structure of H7 hemagglutinin from A/Shanghai2/2013 H7N9 in complex with a human neutralizing antibody H7.HK2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, H7.HK2 Neutralizing Antibody Heavy Chain, H7.HK2 Neutralizing Antibody Light Chain, ... | | Authors: | Morano, N.C, Becker, J.E, Wu, X, Shapiro, L. | | Deposit date: | 2023-08-03 | | Release date: | 2024-05-15 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | CryoEM structure of H7 hemagglutinin from A/Shanghai2/2013 H7N9 in complex with a human neutralizing antibody H7.HK1
To Be Published
|
|
4B4B
 
 | | Pseudomonas aeruginosa RmlA in complex with allosteric inhibitor | | Descriptor: | 1-(2-phenylethyl)pyrimidine-2,4,6(1H,3H,5H)-trione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Alphey, M.S, Pirrie, L, Torrie, L.S, Gardiner, M, Sarkar, A, Brenk, R, Westwood, N.J, Gray, D, Naismith, J.H. | | Deposit date: | 2012-07-30 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Allosteric competitive inhibitors of the glucose-1-phosphate thymidylyltransferase (RmlA) from Pseudomonas aeruginosa.
ACS Chem. Biol., 8, 2013
|
|
3TBN
 
 | |
5XWH
 
 | |
1P0A
 
 | | NMR structure of ETD135, mutant of the antifungal defensin ARD1 from Archaeoprepona demophon | | Descriptor: | DEFENSIN ARD1 | | Authors: | Landon, C, Guenneugues, M, Barbault, F, Legrain, M, Menin, L, Schott, V, Vovelle, F, Dimarcq, J.L. | | Deposit date: | 2003-04-10 | | Release date: | 2004-03-09 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Lead optimization of antifungal peptides with 3D NMR structures analysis.
Protein Sci., 13, 2004
|
|
8AZY
 
 | | KRAS-G12D in complex with BI-2865 | | Descriptor: | (4S)-2-azanyl-4-methyl-4-[3-[4-[(1S)-1-[(2S)-1-methylpyrrolidin-1-ium-2-yl]ethoxy]pyrimidin-2-yl]-1,2,4-oxadiazol-5-yl]-6,7-dihydro-5H-1-benzothiophene-3-carbonitrile, 1,2-ETHANEDIOL, GTPase KRas, ... | | Authors: | Boettcher, J, Herdeis, L. | | Deposit date: | 2022-09-06 | | Release date: | 2023-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Pan-KRAS inhibitor disables oncogenic signalling and tumour growth.
Nature, 619, 2023
|
|
4B6R
 
 | | Structure of Helicobacter pylori Type II Dehydroquinase inhibited by (2S)-2-(4-methoxy)benzyl-3-dehydroquinic acid | | Descriptor: | (1R,2S,4S,5R)-2-(4-methoxyphenyl)methyl-1,4,5-trihydroxy-3-oxocyclohexane-1-carboxylic acid, 3-DEHYDROQUINATE DEHYDRATASE, SULFATE ION | | Authors: | Otero, J.M, Llamas-Saiz, A.L, Lence, E, Tizon, L, Peon, A, Prazeres, V.F.V, Lamb, H, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2012-08-14 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanistic basis of the inhibition of type II dehydroquinase by (2S)- and (2R)-2-benzyl-3-dehydroquinic acids.
ACS Chem. Biol., 8, 2013
|
|
8AZR
 
 | | KRAS in complex with precursor 1 | | Descriptor: | (4~{S})-2-azanyl-4-[3-[6-[(2~{S})-2,4-dimethylpiperazin-1-yl]pyridin-2-yl]-1,2,4-oxadiazol-5-yl]-4-methyl-6,7-dihydro-5~{H}-1-benzothiophene-3-carbonitrile, 1,2-ETHANEDIOL, GTPase KRas, ... | | Authors: | Boettcher, J, Herdeis, L. | | Deposit date: | 2022-09-06 | | Release date: | 2023-06-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Pan-KRAS inhibitor disables oncogenic signalling and tumour growth.
Nature, 619, 2023
|
|
8AZV
 
 | | KRAS in complex with BI-2865 | | Descriptor: | (4S)-2-azanyl-4-methyl-4-[3-[4-[(1S)-1-[(2S)-1-methylpyrrolidin-1-ium-2-yl]ethoxy]pyrimidin-2-yl]-1,2,4-oxadiazol-5-yl]-6,7-dihydro-5H-1-benzothiophene-3-carbonitrile, 1,2-ETHANEDIOL, GTPase KRas, ... | | Authors: | Boettcher, J, Herdeis, L. | | Deposit date: | 2022-09-06 | | Release date: | 2023-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Pan-KRAS inhibitor disables oncogenic signalling and tumour growth.
Nature, 619, 2023
|
|
