5M7P
 
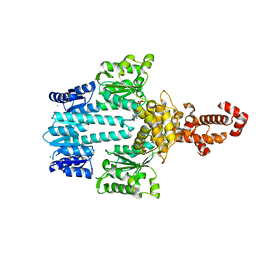 | | Crystal structure of NtrX from Brucella abortus in complex with ADP processed with the CrystalDirect automated mounting and cryo-cooling technology | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Nitrogen assimilation regulatory protein | | Authors: | Cornaciu, I, Fernandez, I, Hoffmann, G, Carrica, M.C, Goldbaum, F.A, Marquez, J.A. | | Deposit date: | 2016-10-28 | | Release date: | 2017-01-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Three-Dimensional Structure of Full-Length NtrX, an Unusual Member of the NtrC Family of Response Regulators.
J. Mol. Biol., 429, 2017
|
|
5MEY
 
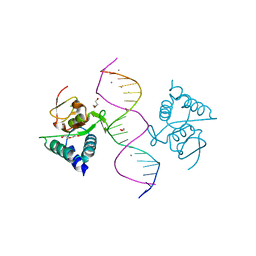 | | Crystal structure of Smad4-MH1 bound to the GGCGC site. | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kaczmarska, Z, Freier, R, Marquez, J.A, Macias, M.J. | | Deposit date: | 2016-11-16 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for genome wide recognition of 5-bp GC motifs by SMAD transcription factors.
Nat Commun, 8, 2017
|
|
5LTV
 
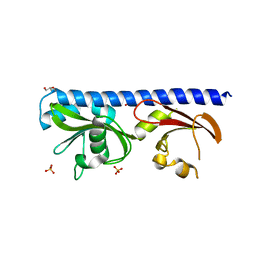 | | LIGAND BINDING DOMAIN OF PSEUDOMONAS AERUGINOSA PAO1 AMINO ACID CHEMORECPETOR PCTC IN COMPLEX WITH GABA | | Descriptor: | ACETATE ION, Chemotactic transducer PctC, GAMMA-AMINO-BUTANOIC ACID, ... | | Authors: | Gavira, J.A, Rico-Jimenez, M, Conejero-Muriel, M, Krell, T. | | Deposit date: | 2016-09-07 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | How Bacterial Chemoreceptors Evolve Novel Ligand Specificities
Mbio, 2020
|
|
8VXY
 
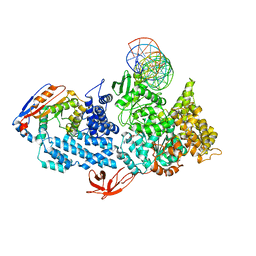 | | Structure of HamA(E138A,K140A)B-plasmid DNA complex from the Escherichia coli Hachiman defense system | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, HamA, HamB, ... | | Authors: | Tuck, O.T, Hu, J.J, Doudna, J.A. | | Deposit date: | 2024-02-06 | | Release date: | 2024-03-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Hachiman is a genome integrity sensor.
Biorxiv, 2024
|
|
8VXA
 
 | |
8VSG
 
 | | SARS-CoV-2 main protease with covalent inhibitor | | Descriptor: | (1R,2S,5S)-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-(1-phenylcyclopropane-1-carbonyl)-3-azabicyclo[3.1.0]hexane-2-carboxamide, 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Bell, J.A, Bandera, A.M. | | Deposit date: | 2024-01-24 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.071 Å) | | Cite: | Exploiting high-energy hydration sites for the discovery of potent peptide aldehyde inhibitors of the SARS-CoV-2 main protease with cellular antiviral activity.
Bioorg.Med.Chem., 103, 2024
|
|
8VXC
 
 | |
5LR8
 
 | | Structure of plastidial phosphorylase Pho1 from Barley | | Descriptor: | Alpha-1,4 glucan phosphorylase, CITRIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Cuesta-Seijo, J.A, Ruzanski, C, Kruzewicz, K, Palcic, M.M. | | Deposit date: | 2016-08-18 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Functional and structural characterization of plastidic starch phosphorylase during barley endosperm development.
PLoS ONE, 12, 2017
|
|
8VX9
 
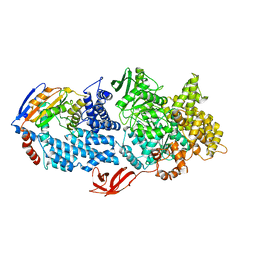 | |
5M1A
 
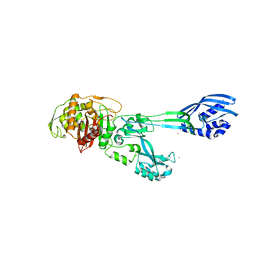 | | Crystal structure of PBP2a from MRSA in the presence of Ceftazidime ligand | | Descriptor: | CADMIUM ION, Penicillin-binding protein 2, beta-muramic acid | | Authors: | Molina, R, Batuecas, M.T, Hermoso, J.A. | | Deposit date: | 2016-10-07 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational Dynamics in Penicillin-Binding Protein 2a of Methicillin-Resistant Staphylococcus aureus, Allosteric Communication Network and Enablement of Catalysis.
J. Am. Chem. Soc., 139, 2017
|
|
5LTX
 
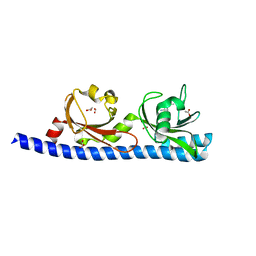 | | LIGAND BINDING DOMAIN OF PSEUDOMONAS AERUGINOSA PAO1 AMINO ACID CHEMORECEPTOR PCTA IN COMPLEX WITH L-MET | | Descriptor: | ACETATE ION, Chemotaxis protein, FORMIC ACID, ... | | Authors: | Gavira, J.A, Rico-Gimenez, M, Ortega, A, Conejero-Muriel, M, Zhulin, I, Krell, T. | | Deposit date: | 2016-09-07 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | How Bacterial Chemoreceptors Evolve Novel Ligand Specificities
Mbio, 2020
|
|
5LWX
 
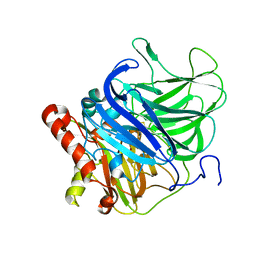 | | Crystal structure of the H253D mutant of McoG from Aspergillus niger | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, ... | | Authors: | Ferraroni, M, Briganti, F, Tamayo-Ramos, J.A, van Berkel, W.J.H, Westphal, A.H. | | Deposit date: | 2016-09-19 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structure and function of Aspergillus niger laccase McoG
Biocatalysis, 2017
|
|
8VV3
 
 | |
5MQZ
 
 | | Archaeal branched-chain amino acid aminotransferase from Archaeoglobus fulgidus; holoform | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | James, P, Isupov, M.N, Sayer, C, Littlechild, J.A, Sutter, J.M, Schmidt, M, Schoenheit, P. | | Deposit date: | 2016-12-21 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Thermostable Branched-Chain Amino Acid Transaminases From the Archaea Geoglobus acetivorans and Archaeoglobus fulgidus : Biochemical and Structural Characterization.
Front Bioeng Biotechnol, 7, 2019
|
|
5MMS
 
 | | Human cystathionine beta-synthase (CBS) p.P49L delta409-551 variant | | Descriptor: | Cystathionine beta-synthase, PROTOPORPHYRIN IX CONTAINING FE, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Vicente, J.B, Colaco, H.G, Malagrino, F, Santo, P.E, Gutierres, A, Bandeiras, T.M, Leandro, P, Brito, J.A, Giuffre, A. | | Deposit date: | 2016-12-12 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Clinically Relevant Variant of the Human Hydrogen Sulfide-Synthesizing Enzyme Cystathionine beta-Synthase: Increased CO Reactivity as a Novel Molecular Mechanism of Pathogenicity?
Oxid Med Cell Longev, 2017, 2017
|
|
5NFQ
 
 | | Novel epoxide hydrolases belonging to the alpha/beta hydrolases superfamily in metagenomes from hot environments | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, IMIDAZOLE, ... | | Authors: | Ferrandi, E.E, De Rose, S.A, Sayer, C, Guazzelli, E, Marchesi, C, Saneei, V, Isupov, M.N, Littlechild, J.A, Monti, D. | | Deposit date: | 2017-03-15 | | Release date: | 2018-05-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | New Thermophilic alpha / beta Class Epoxide Hydrolases Found in Metagenomes From Hot Environments.
Front Bioeng Biotechnol, 6, 2018
|
|
8W1N
 
 | |
8W2W
 
 | |
5NBV
 
 | |
5NM9
 
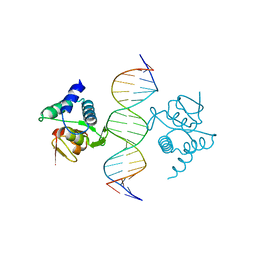 | | Crystal structure of the placozoa Trichoplax adhaerens Smad4-MH1 bound to the GGCGC site. | | Descriptor: | DNA (5'-D(P*AP*TP*GP*CP*GP*GP*GP*CP*GP*CP*GP*CP*CP*CP*GP*CP*AP*T)-3'), Mothers against decapentaplegic homolog, ZINC ION | | Authors: | Kaczmarska, Z, Freier, R, Marquez, J.A, Macias, M.J. | | Deposit date: | 2017-04-05 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural basis for genome wide recognition of 5-bp GC motifs by SMAD transcription factors.
Nat Commun, 8, 2017
|
|
8UZA
 
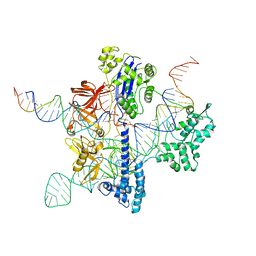 | | Cryo-EM structure of GeoCas9 in complex with sgRNA and target DNA | | Descriptor: | CRISPR-associated endonuclease Cas9, Non-target strand DNA, Target strand DNA, ... | | Authors: | Eggers, A.R, Soczek, K.M, Tuck, O.T, Doudna, J.A. | | Deposit date: | 2023-11-14 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Rapid DNA unwinding accelerates genome editing by engineered CRISPR-Cas9.
Cell, 187, 2024
|
|
5LKX
 
 | | Crystal structure of the p300 acetyltransferase catalytic core with propionyl-coenzyme A. | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, Histone acetyltransferase p300,Histone acetyltransferase p300, ... | | Authors: | Kaczmarska, Z, Ortega, E, Marquez, J.A, Panne, D. | | Deposit date: | 2016-07-25 | | Release date: | 2016-11-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structure of p300 in complex with acyl-CoA variants.
Nat. Chem. Biol., 13, 2017
|
|
8V11
 
 | |
8V10
 
 | |
5M18
 
 | | Crystal structure of PBP2a from MRSA in the presence of Cefepime ligand | | Descriptor: | CADMIUM ION, Penicillin-binding protein 2, beta-muramic acid | | Authors: | Molina, R, Batuecas, M.T, Hermoso, J.A. | | Deposit date: | 2016-10-07 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Conformational Dynamics in Penicillin-Binding Protein 2a of Methicillin-Resistant Staphylococcus aureus, Allosteric Communication Network and Enablement of Catalysis.
J. Am. Chem. Soc., 139, 2017
|
|
