8INP
 
 | | A reversible glycosyltransferase of tectorigenin - Bc7OUGT | | Descriptor: | Bc7OUGT, URIDINE-5'-DIPHOSPHATE, beta-D-glucopyranose | | Authors: | Zhang, Z.Y, Lu, L, Guan, Z.F, Cheng, W.J. | | Deposit date: | 2023-03-10 | | Release date: | 2023-08-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Functional characterization and structural basis of a reversible glycosyltransferase involves in plant chemical defence.
Plant Biotechnol J, 21, 2023
|
|
8ITA
 
 | | A reversible glycosyltransferase of tectorigenin - Bc7OUGT complexed with UDP and tectorigenin | | Descriptor: | 3-(4-hydroxyphenyl)-6-methoxy-5,7-bis(oxidanyl)chromen-4-one, Bc7OUGT, URIDINE-5'-DIPHOSPHATE | | Authors: | Zhang, Z.Y, Lu, L, Guan, Z.F, Cheng, W.J. | | Deposit date: | 2023-03-22 | | Release date: | 2023-08-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Functional characterization and structural basis of a reversible glycosyltransferase involves in plant chemical defence.
Plant Biotechnol J, 21, 2023
|
|
9LKX
 
 | | local refinement of FEM1B bound with PLD6 | | Descriptor: | Mitochondrial cardiolipin hydrolase, Protein fem-1 homolog B | | Authors: | Zhao, S, Xu, C. | | Deposit date: | 2025-01-16 | | Release date: | 2025-04-09 | | Last modified: | 2025-05-07 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | TOM20-driven E3 ligase recruitment regulates mitochondrial dynamics through PLD6.
Nat.Chem.Biol., 2025
|
|
9LKY
 
 | | Dimer of CRL2-FEM1B bound with PLD6 | | Descriptor: | Cullin-2, E3 ubiquitin-protein ligase RBX1, N-terminally processed, ... | | Authors: | Zhao, S, Xu, C. | | Deposit date: | 2025-01-17 | | Release date: | 2025-04-09 | | Last modified: | 2025-05-07 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | TOM20-driven E3 ligase recruitment regulates mitochondrial dynamics through PLD6.
Nat.Chem.Biol., 2025
|
|
9N12
 
 | | Cryo-EM structure of HCoV-HKU1 glycoprotein (mutant T31VPR34 to GGGG) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Jin, M, Rini, J.M. | | Deposit date: | 2025-01-24 | | Release date: | 2025-05-14 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Human coronavirus HKU1 spike structures reveal the basis for sialoglycan specificity and carbohydrate-promoted conformational changes.
Nat Commun, 16, 2025
|
|
9N18
 
 | | Cryo-EM structure of HCoV-HKU1 spike glycoprotein in complex with 9OAc-GD3 tetrasacchride (Deletion 33Pro34Arg) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-acetamido-8-O-(5-acetamido-9-O-acetyl-3,5-dideoxy-D-glycero-alpha-D-galacto-non-2-ulopyranonosyl)-3,5-dideoxy-D-glycero-alpha-D-galacto-non-2-ulopyranosonic acid, ... | | Authors: | Jin, M, Rini, J.M. | | Deposit date: | 2025-01-24 | | Release date: | 2025-05-14 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Human coronavirus HKU1 spike structures reveal the basis for sialoglycan specificity and carbohydrate-promoted conformational changes.
Nat Commun, 16, 2025
|
|
9N17
 
 | | Cryo-EM structure of HCoV-HKU1 glycoprotein D1 Domain (Deletion 33Pro34Arg) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Jin, M, Rini, J.M. | | Deposit date: | 2025-01-24 | | Release date: | 2025-05-14 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Human coronavirus HKU1 spike structures reveal the basis for sialoglycan specificity and carbohydrate-promoted conformational changes.
Nat Commun, 16, 2025
|
|
9N19
 
 | | Cryo-EM structure of HCoV-HKU1spike glycoprotein D1 Domain in complex with 9OAc-GD3 tetrasacchride (Deletion 33Pro34Arg) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-acetamido-8-O-(5-acetamido-9-O-acetyl-3,5-dideoxy-D-glycero-alpha-D-galacto-non-2-ulopyranonosyl)-3,5-dideoxy-D-glycero-alpha-D-galacto-non-2-ulopyranosonic acid, ... | | Authors: | Jin, M, Rini, J.M. | | Deposit date: | 2025-01-24 | | Release date: | 2025-05-14 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Human coronavirus HKU1 spike structures reveal the basis for sialoglycan specificity and carbohydrate-promoted conformational changes.
Nat Commun, 16, 2025
|
|
7F7G
 
 | | a linear Peptide Inhibitors in complex with GK domain | | Descriptor: | DLG4 GK domain, UNK-ARG-ILE-ARG-ARG-ASP-GLU-TYR-LEU-LYS-ALA-ILE-GLN-UNK | | Authors: | Shang, Y, Huang, X, Li, X, Zhang, M. | | Deposit date: | 2021-06-29 | | Release date: | 2022-02-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.446 Å) | | Cite: | Entropy of stapled peptide inhibitors in free state is the major contributor to the improvement of binding affinity with the GK domain.
Rsc Chem Biol, 2, 2021
|
|
9N13
 
 | | Cryo-EM structure of HCoV-HKU1 glycoprotein D1 Domain (mutant T31VPR34 to GGGG) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Jin, M, Rini, J.M. | | Deposit date: | 2025-01-24 | | Release date: | 2025-05-14 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Human coronavirus HKU1 spike structures reveal the basis for sialoglycan specificity and carbohydrate-promoted conformational changes.
Nat Commun, 16, 2025
|
|
9N15
 
 | | Cryo-EM structure of HCoV-HKU1 glycoprotein D1 Domain in complex with 9OAc-GD3(mutant T31VPR34 to GGGG) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-acetamido-8-O-(5-acetamido-9-O-acetyl-3,5-dideoxy-D-glycero-alpha-D-galacto-non-2-ulopyranonosyl)-3,5-dideoxy-D-glycero-alpha-D-galacto-non-2-ulopyranosonic acid, ... | | Authors: | Jin, M, Rini, J.M. | | Deposit date: | 2025-01-24 | | Release date: | 2025-05-14 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Human coronavirus HKU1 spike structures reveal the basis for sialoglycan specificity and carbohydrate-promoted conformational changes.
Nat Commun, 16, 2025
|
|
9N14
 
 | | Cryo-EM structure of HCoV-HKU1 glycoprotein in complex with 9OAc-GD3(mutant T31VPR34 to GGGG) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-acetamido-8-O-(5-acetamido-9-O-acetyl-3,5-dideoxy-D-glycero-alpha-D-galacto-non-2-ulopyranonosyl)-3,5-dideoxy-D-glycero-alpha-D-galacto-non-2-ulopyranosonic acid, ... | | Authors: | Jin, M, Rini, J.M. | | Deposit date: | 2025-01-24 | | Release date: | 2025-05-14 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Human coronavirus HKU1 spike structures reveal the basis for sialoglycan specificity and carbohydrate-promoted conformational changes.
Nat Commun, 16, 2025
|
|
9N16
 
 | | Cryo-EM structure of HCoV-HKU1 glycoprotein (Deletion 33Pro34Arg) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Jin, M, Rini, J.M. | | Deposit date: | 2025-01-24 | | Release date: | 2025-05-14 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Human coronavirus HKU1 spike structures reveal the basis for sialoglycan specificity and carbohydrate-promoted conformational changes.
Nat Commun, 16, 2025
|
|
7F7I
 
 | | Stapled Peptide Inhibitor in complex with PSD95 GK domain | | Descriptor: | ACE-ARG-ILE-ARG-ARG-ASP-GLU-TYR-LEU-LYZ-ALA-ILE-GLN-NH2, Disks large homolog 4 | | Authors: | Shang, Y, Huang, X, Li, X, Zhang, M. | | Deposit date: | 2021-06-29 | | Release date: | 2022-04-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.595 Å) | | Cite: | Entropy of stapled peptide inhibitors in free state is the major contributor to the improvement of binding affinity with the GK domain.
Rsc Chem Biol, 2, 2021
|
|
4YVO
 
 | |
4YCU
 
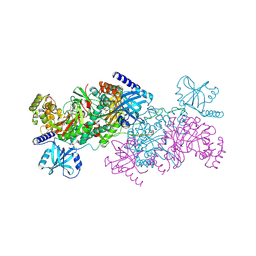 | | Crystal structure of cladosporin in complex with human lysyl-tRNA synthetase | | Descriptor: | Aminoacyl tRNA synthase complex-interacting multifunctional protein 2, GLYCEROL, LYSINE, ... | | Authors: | Fang, P, Wang, J, Guo, M. | | Deposit date: | 2015-02-20 | | Release date: | 2015-06-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Specific Inhibition of tRNA Synthetase by an ATP Competitive Inhibitor.
Chem. Biol., 22, 2015
|
|
4YCV
 
 | |
4YCW
 
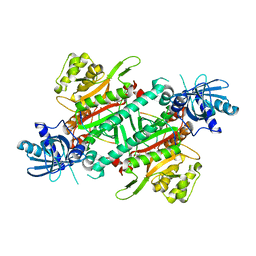 | | Crystal structure of cladosporin in complex with plasmodium like human lysyl-tRNA synthetase mutant | | Descriptor: | Aminoacyl tRNA synthase complex-interacting multifunctional protein 2, LYSINE, Lysine--tRNA ligase, ... | | Authors: | Fang, P, Wang, J, Guo, M. | | Deposit date: | 2015-02-20 | | Release date: | 2015-06-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Basis for Specific Inhibition of tRNA Synthetase by ATP Competitive Inhibitor
Chem.Biol., 22, 2015
|
|
4YVQ
 
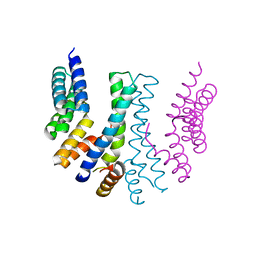 | |
7F46
 
 | | Cryo-EM structure of the SARS-CoV-2 S-6P in complex with 35B5 Fab (state1, local refinement of the RBD, NTD and 35B5 Fab) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 35B5 Fab, Light chain of 35B5 Fab, ... | | Authors: | Wang, X.F, Zhu, Y.Q. | | Deposit date: | 2021-06-17 | | Release date: | 2022-03-23 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.79 Å) | | Cite: | A potent human monoclonal antibody with pan-neutralizing activities directly dislocates S trimer of SARS-CoV-2 through binding both up and down forms of RBD
Signal Transduct Target Ther, 7, 2022
|
|
4HBL
 
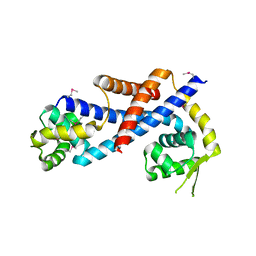 | | Crystal structure of AbfR of Staphylococcus epidermidis | | Descriptor: | Transcriptional regulator, MarR family | | Authors: | Liu, X, Sun, X, Gan, J, Lan, L, Yang, C.-G. | | Deposit date: | 2012-09-28 | | Release date: | 2013-01-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Oxidation-sensing Regulator AbfR Regulates Oxidative Stress Responses, Bacterial Aggregation, and Biofilm Formation in Staphylococcus epidermidis.
J.Biol.Chem., 288, 2013
|
|
4IU6
 
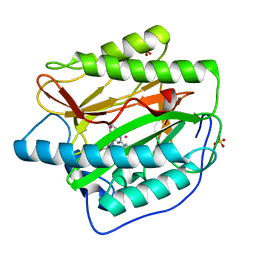 | | Human Methionine Aminopeptidase in complex with FZ1: Pyridinylquinazolines Selectively Inhibit Human Methionine Aminopeptidase-1 | | Descriptor: | 4-[4-(4-methoxyphenyl)piperazin-1-yl]-2-(pyridin-2-yl)quinazoline, COBALT (II) ION, Methionine aminopeptidase 1, ... | | Authors: | Gabelli, S.B, Zhang, F, Miller, M, Liu, J, Amzel, L.M. | | Deposit date: | 2013-01-19 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Pyridinylquinazolines selectively inhibit human methionine aminopeptidase-1 in cells.
J.Med.Chem., 56, 2013
|
|
7R0N
 
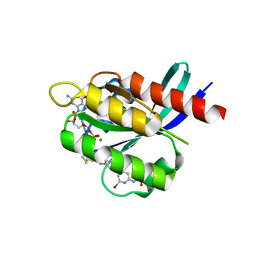 | | KRasG12C in complex with GDP and compound 2 | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ostermann, N. | | Deposit date: | 2022-02-02 | | Release date: | 2022-04-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Discovery, Preclinical Characterization, and Early Clinical Activity of JDQ443, a Structurally Novel, Potent, and Selective Covalent Oral Inhibitor of KRASG12C.
Cancer Discov, 12, 2022
|
|
7R0M
 
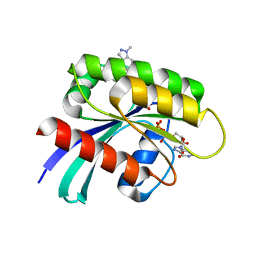 | | KRasG12C in complex with GDP and JDQ443 | | Descriptor: | 1-[6-[4-(5-chloranyl-6-methyl-1~{H}-indazol-4-yl)-5-methyl-3-(1-methylindazol-5-yl)pyrazol-1-yl]-2-azaspiro[3.3]heptan-2-yl]propan-1-one, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Ostermann, N. | | Deposit date: | 2022-02-02 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.611 Å) | | Cite: | Discovery, Preclinical Characterization, and Early Clinical Activity of JDQ443, a Structurally Novel, Potent, and Selective Covalent Oral Inhibitor of KRASG12C.
Cancer Discov, 12, 2022
|
|
7R0Q
 
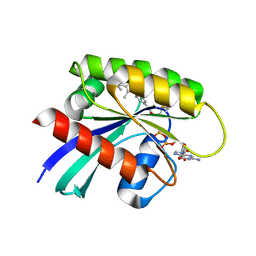 | | KRasG12C in complex with GDP and compound 3 | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ostermann, N. | | Deposit date: | 2022-02-02 | | Release date: | 2022-04-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery, Preclinical Characterization, and Early Clinical Activity of JDQ443, a Structurally Novel, Potent, and Selective Covalent Oral Inhibitor of KRASG12C.
Cancer Discov, 12, 2022
|
|
