6EJG
 
 | |
9JSB
 
 | | guide-bound NbaSPARDA complexes | | Descriptor: | Ago, DREN-APAZ, MAGNESIUM ION, ... | | Authors: | Zhuang, L. | | Deposit date: | 2024-09-30 | | Release date: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Target DNA-induced filament formation and nuclease activation of SPARDA complex.
Cell Res., 2025
|
|
6E7G
 
 | |
9JSP
 
 | | inactive NbaSPARDA complexes | | Descriptor: | Ago, DNA (5'-D(P*GP*CP*TP*GP*TP*GP*CP*AP*GP*TP*AP*TP*T)-3'), DREN-APAZ, ... | | Authors: | Zhuang, L. | | Deposit date: | 2024-09-30 | | Release date: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Target DNA-induced filament formation and nuclease activation of SPARDA complex.
Cell Res., 2025
|
|
5CL0
 
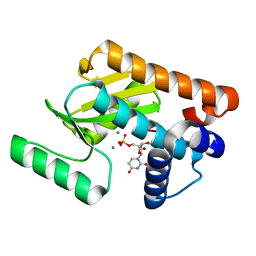 | | 2009 H1N1 PA endonuclease in complex with rUMP | | Descriptor: | MANGANESE (II) ION, Polymerase acidic protein, URIDINE-5'-MONOPHOSPHATE | | Authors: | Kumar, G, White, S.W. | | Deposit date: | 2015-07-15 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Identification and characterization of influenza variants resistant to a viral endonuclease inhibitor.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
9JT2
 
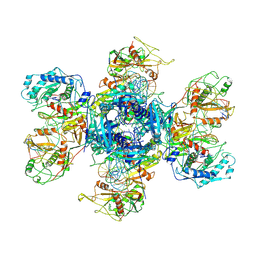 | | substrate-bound NbaSPARDA complexes | | Descriptor: | Ago, DNA (5'-D(*TP*AP*TP*CP*GP*TP*CP*AP*GP*CP*TP*GP*TP*GP*CP*AP*GP*TP*AP*TP*T)-3'), DNA (5'-D(P*GP*AP*TP*AP*CP*TP*AP*C)-3'), ... | | Authors: | Zhuang, L. | | Deposit date: | 2024-10-01 | | Release date: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Target DNA-induced filament formation and nuclease activation of SPARDA complex.
Cell Res., 2025
|
|
9JSZ
 
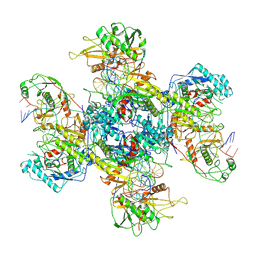 | | active NbaSPARDA complexes | | Descriptor: | Ago, DNA (5'-D(*TP*AP*TP*CP*GP*TP*CP*AP*GP*CP*TP*GP*TP*GP*CP*AP*GP*TP*AP*TP*T)-3'), DREN-APAZ, ... | | Authors: | Zhuang, L. | | Deposit date: | 2024-10-01 | | Release date: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Target DNA-induced filament formation and nuclease activation of SPARDA complex.
Cell Res., 2025
|
|
5CGV
 
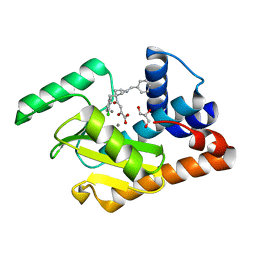 | | 2009 H1N1 PA endonuclease in complex with L-742,001 | | Descriptor: | (2Z)-4-[1-benzyl-4-(4-chlorobenzyl)piperidin-4-yl]-2-hydroxy-4-oxobut-2-enoic acid, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Kumar, G, White, S.W. | | Deposit date: | 2015-07-09 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Identification and characterization of influenza variants resistant to a viral endonuclease inhibitor.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
6EKH
 
 | |
6EJM
 
 | |
6E7I
 
 | | Human ppGalNAcT2 I253A/L310A Mutant with EA2 and UDP | | Descriptor: | EA2, MANGANESE (II) ION, Polypeptide N-acetylgalactosaminyltransferase 2, ... | | Authors: | Bertozzi, C.R, Schumann, B, Agbay, A.J. | | Deposit date: | 2018-07-26 | | Release date: | 2020-01-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bump-and-Hole Engineering Identifies Specific Substrates of Glycosyltransferases in Living Cells.
Mol.Cell, 78, 2020
|
|
3H91
 
 | | Crystal structure of the complex of human chromobox homolog 2 (CBX2) and H3K27 peptide | | Descriptor: | Chromobox protein homolog 2, H3K27 peptide | | Authors: | Amaya, M.F, Ravichandran, M, Loppnau, P, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Min, J, Ouyang, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-04-29 | | Release date: | 2009-08-18 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Recognition and specificity determinants of the human cbx chromodomains.
J.Biol.Chem., 286, 2011
|
|
6EK2
 
 | |
6EKG
 
 | |
6K7P
 
 | | Crystal structure of human AFF4-THD domain | | Descriptor: | AF4/FMR2 family member 4 | | Authors: | Tang, D, Xue, Y, Li, S, Cheng, W, Duan, J, Wang, J, Qi, S. | | Deposit date: | 2019-06-08 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional insight into the effect of AFF4 dimerization on activation of HIV-1 proviral transcription.
Cell Discov, 6, 2020
|
|
5CCU
 
 | | Crystal structure of endoglycoceramidase I from Rhodococ-cus equi | | Descriptor: | 1,2-ETHANEDIOL, Putative secreted endoglycosylceramidase, SODIUM ION | | Authors: | Chen, L. | | Deposit date: | 2015-07-02 | | Release date: | 2015-09-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural Insights into the Broad Substrate Specificity of a Novel Endoglycoceramidase I Belonging to a New Subfamily of GH5 Glycosidases
J. Biol. Chem., 292, 2017
|
|
5D2O
 
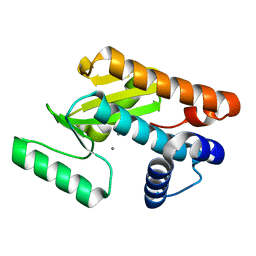 | | 2009 H1N1 PA endonuclease mutant F105S | | Descriptor: | MANGANESE (II) ION, Polymerase acidic protein | | Authors: | Kumar, G, White, S.W. | | Deposit date: | 2015-08-05 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Identification and characterization of influenza variants resistant to a viral endonuclease inhibitor.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3JTY
 
 | | Crystal structure of a BenF-like porin from Pseudomonas fluorescens Pf-5 | | Descriptor: | BenF-like porin, LAURYL DIMETHYLAMINE-N-OXIDE | | Authors: | Sampathkumar, P, Lu, F, Zhao, X, Wasserman, S, Iuzuka, M, Bain, K, Rutter, M, Gheyi, T, Atwell, S, Luz, J, Gilmore, J, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-09-14 | | Release date: | 2009-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structure of a putative BenF-like porin from Pseudomonas fluorescens Pf-5 at 2.6 A resolution.
Proteins, 78, 2010
|
|
8FHS
 
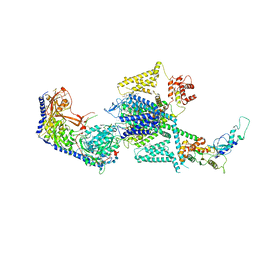 | | Human L-type voltage-gated calcium channel Cav1.2 in the presence of amiodarone and sofosbuvir at 3.3 Angstrom resolution | | Descriptor: | (2-butyl-1-benzofuran-3-yl){4-[2-(diethylamino)ethoxy]-3,5-diiodophenyl}methanone, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gao, S, Yao, X, Yan, N. | | Deposit date: | 2022-12-15 | | Release date: | 2023-12-13 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for human Ca v 1.2 inhibition by multiple drugs and the neurotoxin calciseptine.
Cell, 186, 2023
|
|
6IMC
 
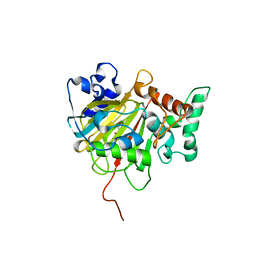 | | Crystal Structure of ALKBH1 in complex with Mn(II) and N-Oxalylglycine | | Descriptor: | MANGANESE (II) ION, N-OXALYLGLYCINE, Nucleic acid dioxygenase ALKBH1 | | Authors: | Zhang, M, Yang, S, Zhao, W, Li, H. | | Deposit date: | 2018-10-22 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Mammalian ALKBH1 serves as an N6-mA demethylase of unpairing DNA.
Cell Res., 30, 2020
|
|
8S9I
 
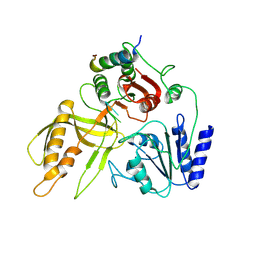 | | Crystal structure of the gp32 C-terminal peptide/Dda/dT8 | | Descriptor: | Dda helicase, dT8, gp32 C-terminal peptide | | Authors: | He, X, Yun, M.K, White, S.W. | | Deposit date: | 2023-03-28 | | Release date: | 2023-06-28 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (3.53 Å) | | Cite: | Structural and functional insights into the interaction between the bacteriophage T4 DNA processing proteins gp32 and Dda.
Nucleic Acids Res., 52, 2024
|
|
8GUN
 
 | |
8GUP
 
 | | Crystal structure of EsaG from Staphylococcus aureus | | Descriptor: | CITRIC ACID, Type VII secretion system protein EsaG | | Authors: | Zhang, Z.M, Wang, Y.J. | | Deposit date: | 2022-09-13 | | Release date: | 2022-11-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.298725 Å) | | Cite: | A toxin-deformation dependent inhibition mechanism in the T7SS toxin-antitoxin system of Gram-positive bacteria.
Nat Commun, 13, 2022
|
|
8GUO
 
 | |
5ZU6
 
 | | A CBM32 derived from alginate lyase B (AlyB-OU02) | | Descriptor: | CBM32 domain, SODIUM ION | | Authors: | Liu, W, Lyu, Q, Zhang, K. | | Deposit date: | 2018-05-07 | | Release date: | 2018-06-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and biochemical characterization of a multidomain alginate lyase reveals a novel role of CBM32 in CAZymes
Biochim. Biophys. Acta, 1862, 2018
|
|
