3HN2
 
 | | Crystal structure of 2-dehydropantoate 2-reductase FROM Geobacter metallireducens GS-15 | | Descriptor: | 2-dehydropantoate 2-reductase | | Authors: | Patskovsky, Y, Toro, R, Morano, C, Rutter, M, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-05-29 | | Release date: | 2009-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of 2-dehydropantoate 2-reductase FROM Geobacter metallireducens
To be Published
|
|
4EJM
 
 | |
3HV2
 
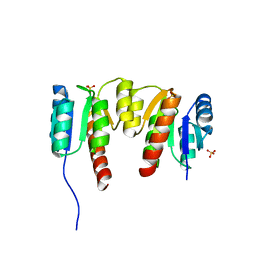 | | Crystal structure of signal receiver domain OF HD domain-containing protein FROM Pseudomonas fluorescens Pf-5 | | Descriptor: | Response regulator/HD domain protein, SULFATE ION | | Authors: | Patskovsky, Y, Ramagopal, U, Toro, R, Freeman, J, Miller, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-06-15 | | Release date: | 2009-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of signal receiver domain oF HD domain-containing protein 3 FROM Pseudomonas fluorescens
To be Published
|
|
4EN0
 
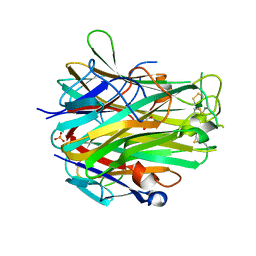 | | Crystal structure of light | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Zhan, C, Liu, W, Patskovsky, Y, Ramagopal, U.A, Bonanno, J.B, Nathenson, S.G, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2012-04-12 | | Release date: | 2012-05-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Mechanistic basis for functional promiscuity in the TNF and TNF receptor superfamilies: structure of the LIGHT:DcR3 assembly.
Structure, 22, 2014
|
|
3I5T
 
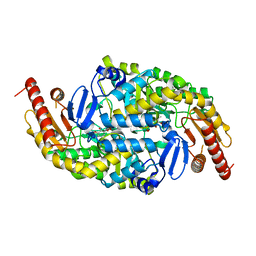 | | CRYSTAL STRUCTURE OF AMINOTRANSFERASE PRK07036 FROM Rhodobacter sphaeroides KD131 | | Descriptor: | Aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Patskovsky, Y, Toro, R, Freeman, J, Do, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-06 | | Release date: | 2009-07-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CRYSTAL STRUCTURE OF AMINOTRANSFERASE PRK07036 FROM Rhodobacter sphaeroides
To be Published
|
|
3HPF
 
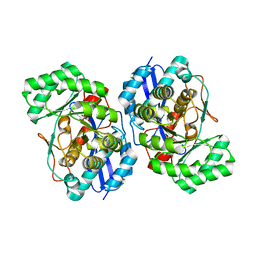 | | Crystal structure of the mutant Y90F of divergent galactarate dehydratase from Oceanobacillus iheyensis complexed with Mg and galactarate | | Descriptor: | D-galactaric acid, MAGNESIUM ION, Muconate cycloisomerase | | Authors: | Fedorov, A.A, Fedorov, E.V, Rakus, J.F, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2009-06-04 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Computation-facilitated assignment of the function in the enolase superfamily: a regiochemically distinct galactarate dehydratase from Oceanobacillus iheyensis .
Biochemistry, 48, 2009
|
|
3I47
 
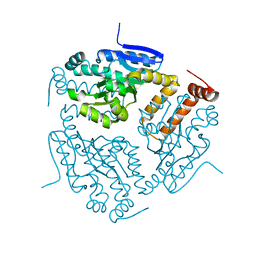 | | CRYSTAL STRUCTURE OF putative enoyl CoA hydratase/isomerase (crotonase) from Legionella pneumophila subsp. pneumophila str. Philadelphia 1 | | Descriptor: | Enoyl CoA hydratase/isomerase (Crotonase) | | Authors: | Malashkevich, V.N, Toro, R, Morano, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-01 | | Release date: | 2009-07-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | CRYSTAL STRUCTURE OF putative enoyl CoA hydratase/isomerase (crotonase) from Legionella pneumophila
subsp. pneumophila str. Philadelphia 1
To be Published
|
|
3HM7
 
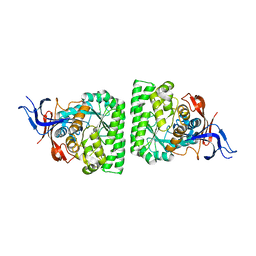 | | Crystal structure of allantoinase from Bacillus halodurans C-125 | | Descriptor: | Allantoinase, ZINC ION | | Authors: | Patskovsky, Y, Romero, R, Rutter, M, Miller, S, Wasserman, S.R, Sauder, J.M, Raushel, F.M, Burley, S.K, Almo, S.C, New York Structural GenomiX Research Consortium (NYSGXRC), New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-05-28 | | Release date: | 2009-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Allantoinase from Bacillus Halodurans
To be Published
|
|
4EZE
 
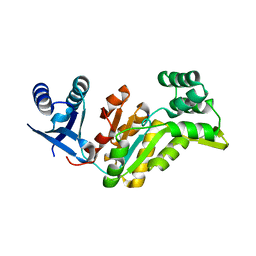 | | Crystal structure of had family hydrolase t0658 from Salmonella enterica subsp. enterica serovar Typhi (Target EFI-501419) | | Descriptor: | CHLORIDE ION, Haloacid dehalogenase-like hydrolase, SODIUM ION | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Allen, K.N, Dunaway-Mariano, D, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-05-02 | | Release date: | 2012-07-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of had hydrolase t0658 from Salmonella enterica (Target EFI-501419)
To be Published
|
|
4F72
 
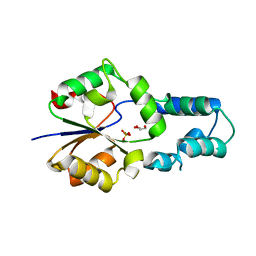 | | Crystal structure of had family enzyme bt-2542 (target efi-501088) from Bacteroides thetaiotaomicron, asp12ala mutant, complex with magnesium and inorganic phosphate | | Descriptor: | GLYCEROL, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Patskovsky, Y, Farelli, J.D, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Zencheck, W.D, Gerlt, J.A, Allen, K.N, Dunaway-Mariano, D, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-05-15 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Protein Bt-2542 from Bacteroides Thetaiotaomicron (Target Efi-501088)
To be Published
|
|
3I6T
 
 | | Crystal structure of muconate cycloisomerase from Jannaschia sp. | | Descriptor: | MAGNESIUM ION, Muconate cycloisomerase, POTASSIUM ION | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Do, J, Romero, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-07 | | Release date: | 2009-07-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of muconate cycloisomerase from Jannaschia sp.
To be Published
|
|
3I0T
 
 | | Sulfur-SAD at long wavelength: Structure of BH3703 from Bacillus halodurans | | Descriptor: | BH3703 protein, SULFATE ION | | Authors: | Ramagopal, U.A, Toro, R, Wasserman, S, Burley, S.K, Almo, S.C. | | Deposit date: | 2009-06-25 | | Release date: | 2009-07-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Sulfur-SAD at long wavelength: Structure of BH3703 from Bacillus halodurans
To be published
|
|
3I45
 
 | | CRYSTAL STRUCTURE OF putative twin-arginine translocation pathway signal protein from Rhodospirillum rubrum Atcc 11170 | | Descriptor: | NICOTINIC ACID, Twin-arginine translocation pathway signal protein | | Authors: | Malashkevich, V.N, Toro, R, Morano, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-01 | | Release date: | 2009-07-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | CRYSTAL STRUCTURE OF putative twin-arginine translocation pathway signal protein from Rhodospirillum rubrum
Atcc 11170
To be Published
|
|
3I6E
 
 | | CRYSTAL STRUCTURE OF MUCONATE LACTONIZING ENZYME FROM Ruegeria pomeroyi. | | Descriptor: | MAGNESIUM ION, Muconate cycloisomerase I, SODIUM ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-07 | | Release date: | 2009-07-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of muconate lactonizing enzyme from Ruegeria pomeroyi.
To be Published
|
|
4F0R
 
 | | Crystal structure of an adenosine deaminase homolog from Chromobacterium violaceum (target NYSGRC-019589) bound Zn and 5'-Methylthioadenosine (unproductive complex) | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, 5-methylthioadenosine/S-adenosylhomocysteine deaminase, GLYCEROL, ... | | Authors: | Kim, J, Vetting, M.W, Sauder, J.M, Burley, S.K, Raushel, F.M, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-05-04 | | Release date: | 2012-06-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of an adenosine deaminase homolog from Chromobacterium violaceum (target NYSGRC-019589) bound Zn and 5'-Methylthioadenosine (unproductive complex)
To be Published
|
|
3IJI
 
 | | Structure of dipeptide epimerase from Bacteroides thetaiotaomicron complexed with L-Ala-D-Glu; nonproductive substrate binding. | | Descriptor: | ALANINE, D-GLUTAMIC ACID, MAGNESIUM ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Lukk, T, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2009-08-04 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3IK4
 
 | | CRYSTAL STRUCTURE OF mandelate racemase/muconate lactonizing protein from Herpetosiphon aurantiacus | | Descriptor: | GLYCEROL, Mandelate racemase/muconate lactonizing protein, POTASSIUM ION | | Authors: | Patskovsky, Y, Toro, R, Dickey, M, Iizuka, M, Sauder, J.M, Gerlt, J.A, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-08-05 | | Release date: | 2009-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4F71
 
 | | Crystal structure of had family enzyme bt-2542 (target efi-501088) from Bacteroides thetaiotaomicron, wild-type protein, complex with magnesium and inorganic phosphate | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Allen, K.N, Dunaway-Mariano, D, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-05-15 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal Structure of Protein Bt-2542 from Bacteroides Thetaiotaomicron (Target Efi-501088)
To be Published
|
|
4F2P
 
 | | Crystal structure of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase from Salmonella enterica with diEtglycol-thio-DADMe-Immucillin-A | | Descriptor: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-({[2-(2-hydroxyethoxy)ethyl]sulfanyl}methyl)pyrrolidin-3- ol, 1,2-ETHANEDIOL, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase, ... | | Authors: | Haapalainen, A.M, Thomas, K, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2012-05-08 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase from Salmonella enterica with diEtglycol-thio-DADMe-Immucillin-A
Structure, 21, 2013
|
|
3IA1
 
 | | Crystal structure of thio-disulfide isomerase from Thermus thermophilus | | Descriptor: | ACETATE ION, Thio-disulfide isomerase/thioredoxin | | Authors: | Patskovsky, Y, Toro, R, Freeman, J, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-13 | | Release date: | 2009-07-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of thio-disulfide isomerase from Thermus thermophilus
To be Published
|
|
3IB6
 
 | | Crystal structure of an uncharacterized protein from Listeria monocytogenes serotype 4b | | Descriptor: | Uncharacterized protein | | Authors: | Bonanno, J.B, Gilmore, M, Bain, K.T, Do, J, Ozyurt, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-15 | | Release date: | 2009-07-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of an uncharacterized protein from Listeria monocytogenes serotype 4b
To be Published
|
|
4EZB
 
 | | CRYSTAL STRUCTURE OF the Conserved hypothetical protein from Sinorhizobium meliloti 1021 | | Descriptor: | uncharacterized conserved protein | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Zenchek, W, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-05-02 | | Release date: | 2012-05-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | CRYSTAL STRUCTURE OF the Conserved hypothetical protein from Sinorhizobium meliloti 1021
To be Published
|
|
4F2D
 
 | | Crystal Structure of Escherichia coli L-arabinose Isomerase (ECAI) complexed with Ribitol | | Descriptor: | ACETIC ACID, D-ribitol, L-arabinose isomerase, ... | | Authors: | Manjasetty, B.A, Burley, S.K, Almo, S.C, Chance, M.R, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2012-05-07 | | Release date: | 2012-05-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Escherichia coli L-arabinose Isomerase (ECAI) complexed with Ribitol
TO BE PUBLISHED
|
|
4DN1
 
 | | Crystal structure of an ENOLASE (mandelate racemase subgroup member) from Agrobacterium tumefaciens (target EFI-502088) with bound mg and formate | | Descriptor: | CHLORIDE ION, FORMIC ACID, Isomerase/lactonizing enzyme, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Bouvier, J.T, Wasserman, S.R, Morisco, L.L, Sojitra, S, Al Obaidi, N.F, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-02-08 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of an enolase (mandelate racemase subgroup member) from Agrobacterium tumefaciens (target EFI-502088) with bound mg and formate
to be published
|
|
4DYE
 
 | | Crystal structure of an enolase (putative sugar isomerase, target efi-502095) from streptomyces coelicolor, no mg, ordered loop | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, isomerase | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-02-28 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of an enolase (putative sugar isomerase, target efi-502095) from streptomyces coelicolor, no mg, ordered loop
to be published
|
|
