3OZ5
 
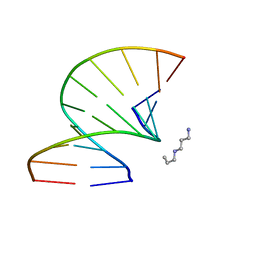 | | S-Methyl Carbocyclic LNA | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*AP*(UMX)P*AP*CP*GP*C)-3'), SPERMINE | | Authors: | Seth, P.R, Allerson, C.A, Berdeja, A, Siwkowski, A, Pallan, P.S, Gaus, H, Prakash, T.P, Watt, A.T, Egli, M, Swayze, E.E. | | Deposit date: | 2010-09-24 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | An exocyclic methylene group acts as a bioisostere of the 2'-oxygen atom in LNA.
J.Am.Chem.Soc., 132, 2010
|
|
4AFJ
 
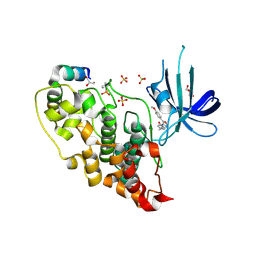 | | 5-aryl-4-carboxamide-1,3-oxazoles: potent and selective GSK-3 inhibitors | | Descriptor: | 5-(4-METHOXYPHENYL)-N-(PYRIDIN-4-YLMETHYL)-1,3-OXAZOLE-4-CARBOXAMIDE, GLYCEROL, GLYCOGEN SYNTHASE KINASE-3 BETA, ... | | Authors: | Gentile, G, Merlo, G, Pozzan, A, Bernasconi, G, Bax, B, Bamborough, P, Bridges, A, Carter, P, Neu, M, Yao, G, Brough, C, Cutler, G, Coffin, A, Belyanskaya, S. | | Deposit date: | 2012-01-19 | | Release date: | 2012-02-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | 5-Aryl-4-Carboxamide-1,3-Oxazoles: Potent and Selective Gsk-3 Inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
1I9S
 
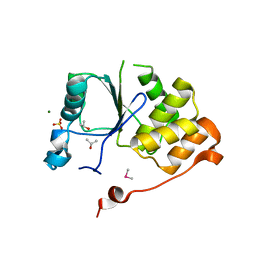 | | CRYSTAL STRUCTURE OF THE RNA TRIPHOSPHATASE DOMAIN OF MOUSE MRNA CAPPING ENZYME | | Descriptor: | CACODYLATE ION, ISOPROPYL ALCOHOL, MAGNESIUM ION, ... | | Authors: | Changela, A, Ho, C.K, Martins, A, Shuman, S, Mondragon, A. | | Deposit date: | 2001-03-20 | | Release date: | 2001-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and mechanism of the RNA triphosphatase component of mammalian mRNA capping enzyme.
EMBO J., 20, 2001
|
|
4L2L
 
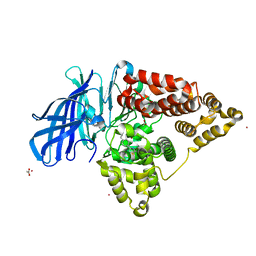 | | Human Leukotriene A4 Hydrolase complexed with ligand 4-(4-benzylphenyl)thiazol-2-amine | | Descriptor: | 4-(4-benzylphenyl)-1,3-thiazol-2-amine, ACETATE ION, Leukotriene A-4 hydrolase, ... | | Authors: | Stsiapanava, A, Rinaldo-Matthis, A, Haeggstrom, J.Z. | | Deposit date: | 2013-06-04 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.648 Å) | | Cite: | Binding of Pro-Gly-Pro at the active site of leukotriene A4 hydrolase/aminopeptidase and development of an epoxide hydrolase selective inhibitor.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4J9D
 
 | |
5H2O
 
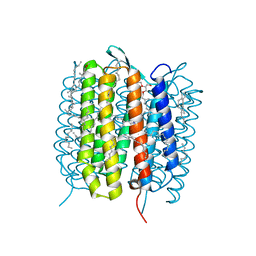 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 250 us after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-10-15 | | Release date: | 2016-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
2YIX
 
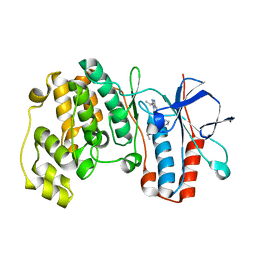 | | Triazolopyridine Inhibitors of p38 | | Descriptor: | 1-ethyl-3-(2-{[3-(1-methylethyl)[1,2,4]triazolo[4,3-a]pyridin-6-yl]sulfanyl}benzyl)urea, MITOGEN-ACTIVATED PROTEIN KINASE 14 | | Authors: | Millan, D.S, Anderson, M, Bunnage, M.E, Burrows, J.L, Butcher, K.J, Dodd, P.G, Evans, T.J, Fairman, D.A, Han, s, Hughes, S.J, Irving, S.L, Kilty, I.C, Lemaitre, A, Lewthawaite, R.A, Mahke, A, Marr, E, Mathias, J.P, Philip, J, Phillips, C, Smith, R.T, Stefaniak, M.H, Yeadon, M. | | Deposit date: | 2011-05-17 | | Release date: | 2011-11-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design and Synthesis of Inhaled P38 Inhibitors for the Treatment of Chronic Obstructive Pulmonary Disease.
J.Med.Chem., 54, 2011
|
|
1I3G
 
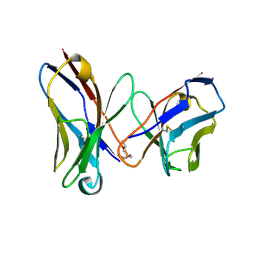 | | CRYSTAL STRUCTURE OF AN AMPICILLIN SINGLE CHAIN FV, FORM 1, FREE | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ANTIBODY FV FRAGMENT | | Authors: | Jung, S, Spinelli, S, Schimmele, B, Honegger, A, Pugliese, L, Cambillau, C, Pluckthun, A. | | Deposit date: | 2001-02-15 | | Release date: | 2001-10-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Selection, characterization and x-ray structure of anti-ampicillin single-chain Fv fragments from phage-displayed murine antibody libraries.
J.Mol.Biol., 309, 2001
|
|
3LOQ
 
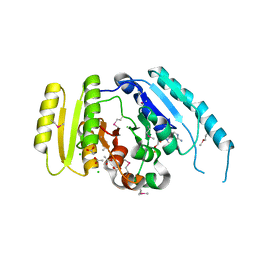 | | The crystal structure of a universal stress protein from Archaeoglobus fulgidus DSM 4304 | | Descriptor: | ACETATE ION, ADENOSINE MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Tan, K, Weger, A, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-02-04 | | Release date: | 2010-02-16 | | Last modified: | 2012-02-22 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | The crystal structure of a universal stress protein from Archaeoglobus fulgidus DSM 4304
To be Published
|
|
3P5X
 
 | | Actinidin from Actinidia arguta planch (Sarusashi) | | Descriptor: | Actinidin, CADMIUM ION | | Authors: | Manickam, Y, Nirmal, N, Suzuki, A, Sugiyama, Y, Yamane, T, Devadasan, V, Sharma, A. | | Deposit date: | 2010-10-11 | | Release date: | 2010-11-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural analysis of actinidin and a comparison of cadmium and sulfur anomalous signals from actinidin crystals measured using in-house copper- and chromium-anode X-ray sources
Acta Crystallogr.,Sect.D, 66, 2010
|
|
5FDS
 
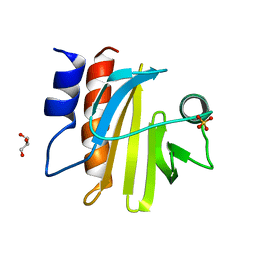 | |
3P52
 
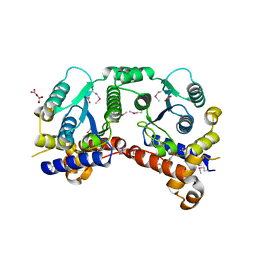 | | NH3-dependent NAD synthetase from Campylobacter jejuni subsp. jejuni NCTC 11168 in complex with the nitrate ion | | Descriptor: | NH(3)-dependent NAD(+) synthetase, NITRATE ION | | Authors: | Filippova, E.V, Wawrzak, Z, Onopriyenko, O, Skarina, T, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-10-07 | | Release date: | 2010-10-27 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | NH3-dependent NAD synthetase from Campylobacter jejuni subsp. jejuni NCTC 11168 in complex with the nitrate ion
To be Published
|
|
4L7W
 
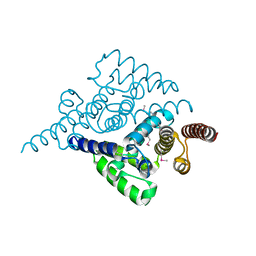 | | Crystal structure mutant H77A of human HD domain-containing protein 2, Genomics Consortium (NESG) Target HR6723 | | Descriptor: | HD domain-containing protein 2, MAGNESIUM ION | | Authors: | Kuzin, A, Su, M, Yakunin, A, Beloglazova, N, Seetharaman, J, Maglaqui, M, Xiao, R, Lee, D, Brown, G, Flick, R, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-06-14 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.305 Å) | | Cite: | Northeast Structural Genomics Consortium Target HR6723
To be Published
|
|
3P5U
 
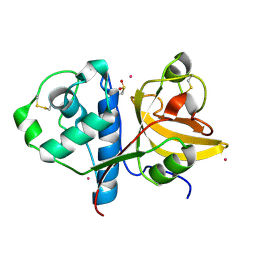 | | Actinidin from Actinidia arguta planch (Sarusashi) | | Descriptor: | Actinidin, CADMIUM ION | | Authors: | Manickam, Y, Nirmal, N, Suzuki, A, Sugiyama, Y, Yamane, T, Devadasan, V, Sharma, A. | | Deposit date: | 2010-10-11 | | Release date: | 2010-11-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural analysis of actinidin and a comparison of cadmium and sulfur anomalous signals from actinidin crystals measured using in-house copper- and chromium-anode X-ray sources
Acta Crystallogr.,Sect.D, 66, 2010
|
|
4L9G
 
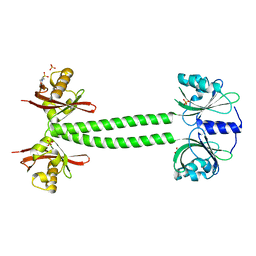 | | Structure of PpsR N-Q-PAS1 from Rb. sphaeroides | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, SULFATE ION, Transcriptional regulator, ... | | Authors: | Heintz, U, Meinhart, A, Schlichting, I, Winkler, A. | | Deposit date: | 2013-06-18 | | Release date: | 2014-02-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Multi-PAS domain-mediated protein oligomerization of PpsR from Rhodobacter sphaeroides.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3P9A
 
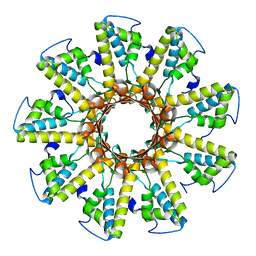 | |
1WDD
 
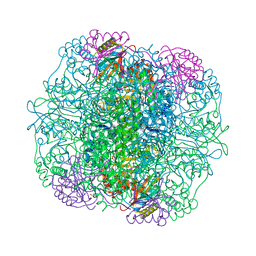 | | Crystal Structure of Activated Rice Rubisco Complexed with 2-Carboxyarabinitol-1,5-bisphosphate | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Mizohata, E, Matsumura, H, Ueno, T, Ishida, H, Inoue, T, Makino, A, Mae, T, Kai, Y. | | Deposit date: | 2004-05-13 | | Release date: | 2004-11-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of rice Rubisco and implications for activation induced by positive effectors NADPH and 6-phosphogluconate
J.Mol.Biol., 422, 2012
|
|
7XXF
 
 | | Structure of photosynthetic LH1-RC super-complex of Rhodopila globiformis | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (6~{E},8~{E},10~{E},12~{E},14~{E},16~{E},18~{E},20~{E},22~{E},24~{E},26~{E},28~{E})-2,31-dimethoxy-2,6,10,14,19,23,27,31-octamethyl-dotriaconta-6,8,10,12,14,16,18,20,22,24,26,28-dodecaen-5-one, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ... | | Authors: | Tani, K, Kanno, R, Kurosawa, K, Takaichi, S, Nagashima, K.V.P, Hall, M, Yu, L.-J, Kimura, Y, Madigan, M.T, Mizoguchi, A, Humbel, B.M, Wang-Otomo, Z.-Y. | | Deposit date: | 2022-05-30 | | Release date: | 2022-11-16 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.24 Å) | | Cite: | An LH1-RC photocomplex from an extremophilic phototroph provides insight into origins of two photosynthesis proteins.
Commun Biol, 5, 2022
|
|
5FP2
 
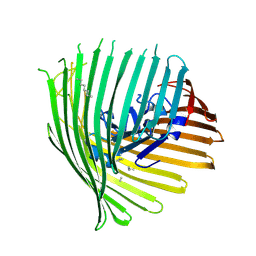 | |
2GFN
 
 | | Crystal structure of HTH-type transcriptional regulator pksA related protein from Rhodococcus sp. RHA1 | | Descriptor: | CHLORIDE ION, HTH-type transcriptional regulator pksA related protein | | Authors: | Nocek, B, Evdokimova, E, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-03-22 | | Release date: | 2006-04-25 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of HTH-type transcriptional regulator pksA related protein from Rhodococcus sp. RHA1
To be Published
|
|
4JJB
 
 | |
4A6V
 
 | | X-ray structures of oxazole hydroxamate EcMetAp-Mn complexes | | Descriptor: | CARBONATE ION, MANGANESE (II) ION, METHIONINE AMINOPEPTIDASE, ... | | Authors: | Huguet, F, Melet, A, AlvesdeSousa, R, Lieutaud, A, Chevalier, J, Deschamps, P, Tomas, A, Leulliot, N, Pages, J.M, Artaud, I. | | Deposit date: | 2011-11-09 | | Release date: | 2012-06-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Hydroxamic Acids as Potent Inhibitors of Fe(II) and Mn(II) E. Coli Methionine Aminopeptidase: Biological Activities and X-Ray Structures of Oxazole Hydroxamate-Ecmetap-Mn Complexes.
Chemmedchem, 7, 2012
|
|
4AHP
 
 | | Crystal Structure of Wild Type N-acetylneuraminic acid lyase from Staphylococcus aureus | | Descriptor: | CHLORIDE ION, N-ACETYLNEURAMINATE LYASE | | Authors: | Timms, N, Polyakova, A, Windle, C.L, Trinh, C.H, Nelson, A, Trinh, A.R, Berry, A. | | Deposit date: | 2012-02-06 | | Release date: | 2013-01-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Insights Into the Recovery of Aldolase Activity in N-Acetylneuraminic Acid Lyase by Replacement of the Catalytically Active Lysine with Gamma-Thialysine by Using a Chemical Mutagenesis Strategy.
Chembiochem, 14, 2013
|
|
3OO9
 
 | |
2GEN
 
 | | Structural Genomics, the crystal structure of a probable transcriptional regulator from Pseudomonas aeruginosa PAO1 | | Descriptor: | probable transcriptional regulator | | Authors: | Tan, K, Skarina, T, Onopriyenko, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-03-20 | | Release date: | 2006-04-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of a probable transcriptional regulator from Pseudomonas aeruginosa PAO1
To be Published
|
|
