1UNW
 
 | | Structure Based Engineering of Internal Molecular Surfaces Of Four Helix Bundles | | Descriptor: | GENERAL CONTROL PROTEIN GCN4 | | Authors: | Yadav, M.K, Redman, J.E, Alvarez-Gutierrez, J.M, Zhang, Y, Stout, C.D, Ghadiri, M.R. | | Deposit date: | 2003-09-15 | | Release date: | 2004-10-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Engineering of Internal Cavities in Coiled-Coil Peptides
Biochemistry, 44, 2005
|
|
1M7R
 
 | | Crystal Structure of Myotubularin-related Protein-2 (MTMR2) Complexed with Phosphate | | Descriptor: | Myotubularin-related Protein-2, PHOSPHATE ION | | Authors: | Begley, M.J, Taylor, G.S, Kim, S.-A, Veine, D.M, Dixon, J.E, Stuckey, J.A. | | Deposit date: | 2002-07-22 | | Release date: | 2003-10-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a phosphoinositide phosphatase, MTMR2: insights into myotubular myopathy and Charcot-Marie-Tooth syndrome
Mol.Cell, 12, 2003
|
|
1MR2
 
 | | Structure of the MT-ADPRase in complex with 1 Mn2+ ion and AMP-CP (a inhibitor), a nudix enzyme | | Descriptor: | ADPR pyrophosphatase, MANGANESE (II) ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER | | Authors: | Kang, L.-W, Gabelli, S.B, Bianchet, M.A, Cunningham, J.E, O'Handley, S.F, Amzel, L.M. | | Deposit date: | 2002-09-17 | | Release date: | 2003-08-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and mechanism of MT-ADPRase, a Nudix hydrolase from Mycobacterium tuberculosis
Structure, 11, 2003
|
|
1MQW
 
 | | Structure of the MT-ADPRase in complex with three Mn2+ ions and AMPCPR, a Nudix enzyme | | Descriptor: | ADPR pyrophosphatase, ALPHA-BETA METHYLENE ADP-RIBOSE, MANGANESE (II) ION | | Authors: | Kang, L.-W, Gabelli, S.B, Bianchet, M.A, Cunningham, J.E, O'Handley, S.F, Amzel, L.M. | | Deposit date: | 2002-09-17 | | Release date: | 2003-08-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and mechanism of MT-ADPRase, a Nudix hydrolase from Mycobacterium tuberculosis
Structure, 11, 2003
|
|
1NF8
 
 | | Crystal structure of PhzD protein active site mutant with substrate | | Descriptor: | (5S,6S)-5-[(1-carboxyethenyl)oxy]-6-hydroxycyclohexa-1,3-diene-1-carboxylic acid, octyl beta-D-glucopyranoside, phenazine biosynthesis protein phzD | | Authors: | Parsons, F, Calabrese, K, Eisenstein, E, Ladner, J.E. | | Deposit date: | 2002-12-13 | | Release date: | 2003-06-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and mechanism of Pseudomonas aeruginosa PhzD, an isochorismatase from the phenazine biosynthetic pathway
Biochemistry, 42, 2003
|
|
1NC1
 
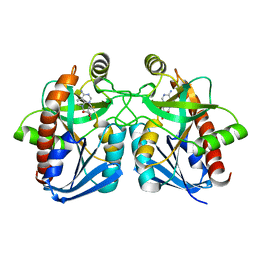 | | Crystal structure of E. coli MTA/AdoHcy nucleosidase complexed with 5'-methylthiotubercidin (MTH) | | Descriptor: | 2-(4-AMINO-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-METHYLSULFANYLMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, MTA/SAH nucleosidase | | Authors: | Lee, J.E, Cornell, K.A, Riscoe, M.K, Howell, P.L. | | Deposit date: | 2002-12-04 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Escherichia coli 5'-methylthioadenosine/ S-adenosylhomocysteine nucleosidase inhibitor complexes provide insight into the conformational changes required for substrate binding and catalysis.
J.Biol.Chem., 278, 2003
|
|
1M7Q
 
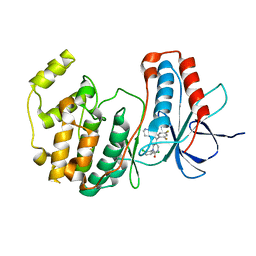 | | Crystal structure of p38 MAP kinase in complex with a dihydroquinazolinone inhibitor | | Descriptor: | 1-(2,6-DICHLOROPHENYL)-5-(2,4-DIFLUOROPHENYL)-7-PIPERAZIN-1-YL-3,4-DIHYDROQUINAZOLIN-2(1H)-ONE, Mitogen-activated protein kinase 14, SULFATE ION | | Authors: | Stelmach, J.E, Liu, L, Patel, S.B, Pivnichny, J.V, Scapin, G, Singh, S, Hop, C.E.C.A, Wang, Z, Cameron, P.M, Nichols, E.A, O'Keefe, S.J, O'Neill, E.A, Schmatz, D.M, Schwartz, C.D, Thompson, C.M, Zaller, D.M, Doherty, J.B. | | Deposit date: | 2002-07-22 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design and synthesis of potent, orally bioavailable dihydroquinazolinone inhibitors of p38 MAP kinase.
Bioorg.Med.Chem.Lett., 13, 2003
|
|
1NH3
 
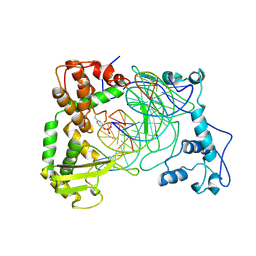 | | Human Topoisomerase I Ara-C Complex | | Descriptor: | 5'-D(*(GNG)P*GP*AP*AP*AP*AP*AP*UP*UP*UP*UP*T)-3', 5'-D(*AP*AP*AP*AP*AP*GP*AP*CP*UP*(UBB))-3', 5'-D(*AP*AP*AP*AP*AP*TP*UP*UP*UP*UP*CP*(CAR)P*AP*AP*GP*UP*CP*UP*UP*UP*UP*T)-3', ... | | Authors: | Chrencik, J.E, Burgin, A.B, Pommier, Y, Stewart, L, Redinbo, M.R. | | Deposit date: | 2002-12-18 | | Release date: | 2003-03-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Impact of the Leukemia Drug 1-beta-D-Arabinofuranosylcytosine (Ara-C) on the Covalent Human Topoisomerase I-DNA Complex
J.Biol.Chem., 278, 2003
|
|
1NF9
 
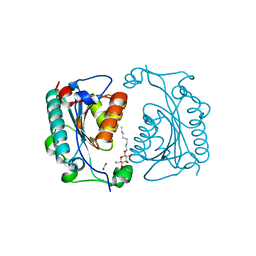 | | Crystal Structure of PhzD protein from Pseudomonas aeruginosa | | Descriptor: | FORMIC ACID, octyl beta-D-glucopyranoside, phenazine biosynthesis protein phzD | | Authors: | Parsons, F, Calabrese, K, Eisenstein, E, Ladner, J.E. | | Deposit date: | 2002-12-13 | | Release date: | 2003-06-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and mechanism of Pseudomonas aeruginosa PhzD, an isochorismatase from the phenazine biosynthetic pathway
Biochemistry, 42, 2003
|
|
1LVB
 
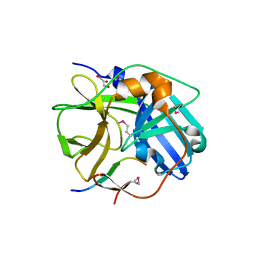 | | CATALYTICALLY INACTIVE TOBACCO ETCH VIRUS PROTEASE COMPLEXED WITH SUBSTRATE | | Descriptor: | CATALYTIC DOMAIN OF THE NUCLEAR INCLUSION PROTEIN A (NIA), GLYCEROL, OLIGOPEPTIDE SUBSTRATE FOR THE PROTEASE | | Authors: | Phan, J, Zdanov, A, Evdokimov, A.G, Tropea, J.E, Peters III, H.K, Kapust, R.B, Li, M, Wlodawer, A, Waugh, D.S. | | Deposit date: | 2002-05-28 | | Release date: | 2002-11-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the substrate specificity of tobacco etch virus protease.
J.Biol.Chem., 277, 2002
|
|
1MAZ
 
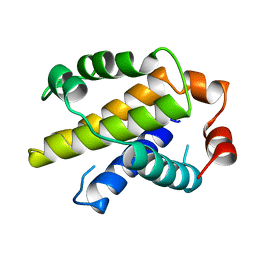 | | X-RAY STRUCTURE OF BCL-XL, AN INHIBITOR OF PROGRAMMED CELL DEATH | | Descriptor: | Bcl-2-like protein 1 | | Authors: | Muchmore, S.W, Sattler, M, Liang, H, Meadows, R.P, Harlan, J.E, Yoon, H.S, Nettesheim, D, Chang, B.S, Thompson, C.B, Wong, S.L, Ng, S.C, Fesik, S.W. | | Deposit date: | 1996-04-09 | | Release date: | 1997-04-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray and NMR structure of human Bcl-xL, an inhibitor of programmed cell death.
Nature, 381, 1996
|
|
7L7E
 
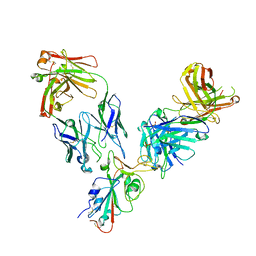 | |
7L7D
 
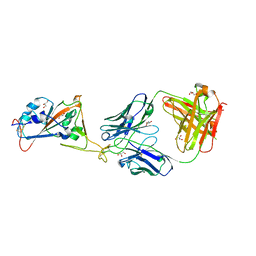 | |
1NC3
 
 | | Crystal structure of E. coli MTA/AdoHcy nucleosidase complexed with formycin A (FMA) | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, MTA/SAH nucleosidase | | Authors: | Lee, J.E, Cornell, K.A, Riscoe, M.K, Howell, P.L. | | Deposit date: | 2002-12-04 | | Release date: | 2003-03-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Escherichia coli 5'-methylthioadenosine/ S-adenosylhomocysteine nucleosidase inhibitor complexes provide insight into the conformational changes required for substrate binding and catalysis.
J.Biol.Chem., 278, 2003
|
|
1X9F
 
 | | Hemoglobin Dodecamer from Lumbricus Erythrocruorin | | Descriptor: | CARBON MONOXIDE, Globin II, extracellular, ... | | Authors: | Strand, K, Knapp, J.E, Bhyravbhatla, B, Royer Jr, W.E. | | Deposit date: | 2004-08-20 | | Release date: | 2004-11-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the hemoglobin dodecamer from lumbricus erythrocruorin: allosteric core of giant annelid respiratory complexes
J.Mol.Biol., 344, 2004
|
|
1X7D
 
 | | Crystal Structure Analysis of Ornithine Cyclodeaminase Complexed with NAD and ornithine to 1.6 Angstroms | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, L-ornithine, ... | | Authors: | Alam, S, Goodman, J.L, Wang, S, Ruzicka, F.J, Frey, P.A, Wedekind, J.E. | | Deposit date: | 2004-08-13 | | Release date: | 2004-11-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Ornithine Cyclodeaminase: Structure, Mechanism of Action, and Implications for
the u-Crystallin Family
Biochemistry, 43, 2004
|
|
1LA1
 
 | | Gro-EL Fragment (Apical Domain) Comprising Residues 188-379 | | Descriptor: | GroEL | | Authors: | Ashcroft, A.E, Brinker, A, Coyle, J.E, Weber, F, Kaiser, M, Moroder, L, Parsons, M.R, Jager, J, Hartl, U.F, Hayer-Hartl, M, Radford, S.E. | | Deposit date: | 2002-03-27 | | Release date: | 2002-04-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural plasticity and noncovalent substrate binding in the GroEL apical domain. A study using electrospay ionization mass spectrometry and fluorescence binding studies.
J.Biol.Chem., 277, 2002
|
|
1NWI
 
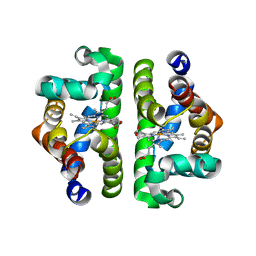 | |
1NMP
 
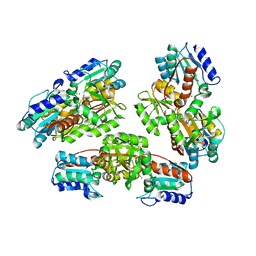 | | Structural genomics, ybgI protein, unknown function | | Descriptor: | Hypothetical protein ybgI, MAGNESIUM ION | | Authors: | Ladner, J.E, Obmolova, G, Teplyakov, A, Khil, P.P, Camerini-Otero, R.D, Gilliland, G.L, Structure 2 Function Project (S2F) | | Deposit date: | 2003-01-10 | | Release date: | 2004-01-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Escherichia coli Protein ybgI, a toroidal structure with a dinuclear metal site
BMC Struct.Biol., 3, 2003
|
|
1VKC
 
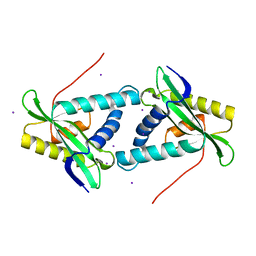 | | Putative acetyl transferase from Pyrococcus furiosus | | Descriptor: | IODIDE ION, putative acetyl transferase | | Authors: | Habel, J.E, Liu, Z.-J, Tempel, W, Rose, J.P, Brereton, P.S, Izumi, M, Jenney Jr, F.E, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Richardson, D.C, Richardson, J.S, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-05-11 | | Release date: | 2004-12-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Putative acetyl transferase from Pyrococcus furiosus
To be published
|
|
1W7Z
 
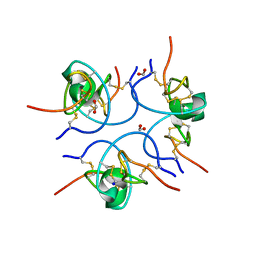 | | Crystal structure of the free (uncomplexed) Ecballium elaterium trypsin inhibitor (EETI-II) | | Descriptor: | FORMIC ACID, SODIUM ION, TRYPSIN INHIBITOR II | | Authors: | Kraetzner, R, Debreczeni, J.E, Pape, T, Kolmar, H, Schneider, T.R, Uson, I, Scheldrick, G.M. | | Deposit date: | 2004-09-14 | | Release date: | 2005-11-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structure of Ecballium Elaterium Trypsin Inhibitor II (Eeti-II): A Rigid Molecular Scaffold
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1UNY
 
 | | Structure Based Engineering of Internal Molecular Surfaces Of Four Helix Bundles | | Descriptor: | GENERAL CONTROL PROTEIN GCN4 | | Authors: | Yadav, M.K, Redman, J.E, Alvarez-Gutierrez, J.M, Zhang, Y, Stout, C.D, Ghadiri, M.R. | | Deposit date: | 2003-09-15 | | Release date: | 2004-10-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Engineering of Internal Cavities in Coiled-Coil Peptides
Biochemistry, 44, 2005
|
|
1NXF
 
 | |
1NWN
 
 | |
1NMO
 
 | | Structural genomics, protein ybgI, unknown function | | Descriptor: | FE (III) ION, Hypothetical protein ybgI | | Authors: | Ladner, J.E, Obmolova, G, Teplyakov, A, Khil, P.P, Camerini-Otero, R.D, Gilliland, G.L, Structure 2 Function Project (S2F) | | Deposit date: | 2003-01-10 | | Release date: | 2004-01-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Escherichia coli Protein ybgI, a toroidal structure with a dinuclear metal site
BMC Struct.Biol., 3, 2003
|
|
