6T06
 
 | | Crystal structure of YTHDC1 with fragment 19 (DHU_DC1_045) | | Descriptor: | 3-imidazolidin-2-yl-2~{H}-indazole, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-10-02 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
6T0C
 
 | | Crystal structure of YTHDC1 with fragment 26 (DHU_DC1_198) | | Descriptor: | SULFATE ION, YTHDC1, ~{N}-methyl-2~{H}-indazole-3-carboxamide | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-10-02 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
6SZ3
 
 | | Crystal structure of YTHDC1 with fragment 4 (DHU_DC1_158) | | Descriptor: | SULFATE ION, YTH domain-containing protein 1, ~{N}-methyl-5,6,7,8-tetrahydroquinolin-4-amine | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-10-01 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
6T03
 
 | | Crystal structure of YTHDC1 with fragment 16 (DHU_DC1_017) | | Descriptor: | 1,3-dihydroimidazole-2-thione, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-10-02 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
6T08
 
 | | Crystal structure of YTHDC1 with fragment 21 (DHU_DC1_131) | | Descriptor: | SULFATE ION, YTHDC1, ~{N}-(1~{H}-imidazol-2-yl)thiophene-2-sulfonamide | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-10-02 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
6SZ2
 
 | | Crystal structure of YTHDC1 with fragment 3 (DHU_DC1_149) | | Descriptor: | SULFATE ION, YTH domain-containing protein 1, ~{N}-methyl-1,6-naphthyridin-4-amine | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-10-01 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
3FDR
 
 | | Crystal structure of TDRD2 | | Descriptor: | Tudor and KH domain-containing protein | | Authors: | Amaya, M.F, Adams, M.A, Guo, Y, Li, Y, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-11-26 | | Release date: | 2009-01-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Mouse Piwi interactome identifies binding mechanism of Tdrkh Tudor domain to arginine methylated Miwi
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
6ILM
 
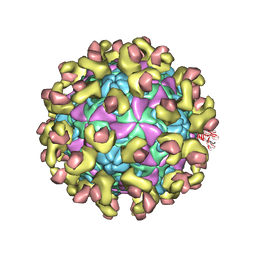 | | Cryo-EM structure of Echovirus 6 complexed with its uncoating receptor FcRn at PH 7.4 | | Descriptor: | Beta-2-microglobulin, Capsid protein VP1, Capsid protein VP2, ... | | Authors: | Gao, G.F, Liu, S, Zhao, X, Peng, R. | | Deposit date: | 2018-10-19 | | Release date: | 2019-05-15 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Human Neonatal Fc Receptor Is the Cellular Uncoating Receptor for Enterovirus B.
Cell, 177, 2019
|
|
6ILO
 
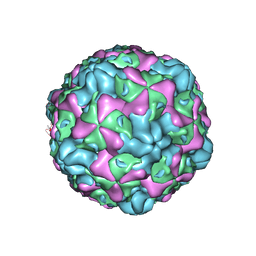 | | Cryo-EM structure of empty Echovirus 6 particle at PH 7.4 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Gao, G.F, Liu, S, Zhao, X, Peng, R. | | Deposit date: | 2018-10-19 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Human Neonatal Fc Receptor Is the Cellular Uncoating Receptor for Enterovirus B.
Cell, 177, 2019
|
|
6ILL
 
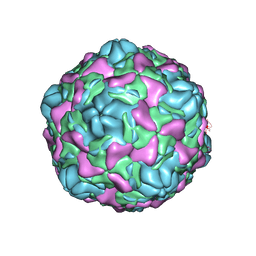 | | Cryo-EM structure of Echovirus 6 complexed with its uncoating receptor FcRn at PH 5.5 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Gao, G.F, Liu, S, Zhao, X, Peng, R. | | Deposit date: | 2018-10-19 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Human Neonatal Fc Receptor Is the Cellular Uncoating Receptor for Enterovirus B.
Cell, 177, 2019
|
|
6ILJ
 
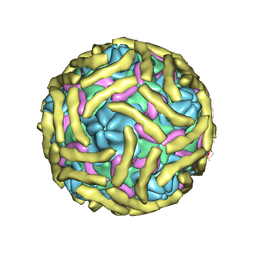 | | Cryo-EM structure of Echovirus 6 complexed with its attachment receptor CD55 at PH 5.5 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Gao, G.F, Liu, S, Zhao, X, Peng, R. | | Deposit date: | 2018-10-18 | | Release date: | 2019-05-15 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Human Neonatal Fc Receptor Is the Cellular Uncoating Receptor for Enterovirus B.
Cell, 177, 2019
|
|
6ILK
 
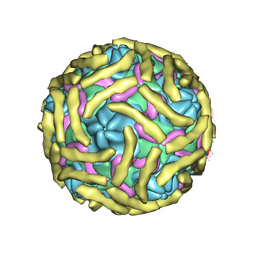 | | Cryo-EM structure of Echovirus 6 complexed with its attachment receptor CD55 at PH 7.4 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Gao, G.F, Liu, S, Zhao, X, Peng, R. | | Deposit date: | 2018-10-18 | | Release date: | 2019-05-15 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Human Neonatal Fc Receptor Is the Cellular Uncoating Receptor for Enterovirus B.
Cell, 177, 2019
|
|
8JOP
 
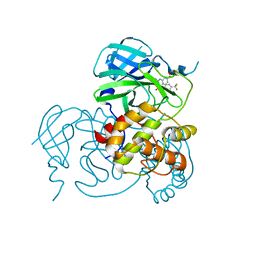 | | Crystal structure of the SARS-CoV-2 main protease in complex with 11a | | Descriptor: | 3C-like proteinase nsp5, methyl (6~{R})-5-ethanoyl-7-oxidanylidene-6-[4-(trifluoromethyl)phenyl]-8,9,10,11-tetrahydro-6~{H}-benzo[b][1,4]benzodiazepine-2-carboxylate | | Authors: | Zeng, R, Liu, Y.Z, Wang, F.L, Yang, S.Y, Lei, J. | | Deposit date: | 2023-06-08 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of benzodiazepine derivatives as a new class of covalent inhibitors of SARS-CoV-2 main protease.
Bioorg.Med.Chem.Lett., 92, 2023
|
|
6IVU
 
 | |
6IVS
 
 | |
8WWW
 
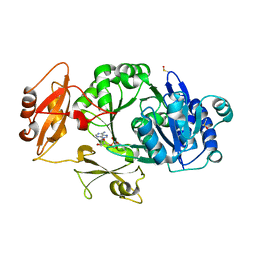 | | Crystal structure of the adenylation domain of SyAstC | | Descriptor: | Carrier domain-containing protein, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Li, Z, Zhu, Z, Zhu, D, Shen, Y. | | Deposit date: | 2023-10-27 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Functional Application of the Single-Module NRPS-like d-Alanyltransferase in Maytansinol Biosynthesis
Acs Catalysis, 14, 2024
|
|
4MNQ
 
 | | TCR-peptide specificity overrides affinity enhancing TCR-MHC interactions | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Rizkallah, P.J, Cole, D.K, Sewell, A.K, Jakobsen, B.K. | | Deposit date: | 2013-09-11 | | Release date: | 2013-11-13 | | Last modified: | 2017-08-02 | | Method: | X-RAY DIFFRACTION (2.742 Å) | | Cite: | T-cell receptor (TCR)-peptide specificity overrides affinity-enhancing TCR-major histocompatibility complex interactions.
J.Biol.Chem., 289, 2014
|
|
8Y3X
 
 | | Cell divisome sPG hydrolysis machinery FtsEX-EnvC | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division ATP-binding protein FtsE, Cell division protein FtsX, ... | | Authors: | Zhang, Z, Dong, H, Chen, Y. | | Deposit date: | 2024-01-29 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Structure and activity of the septal peptidoglycan hydrolysis machinery crucial for bacterial cell division.
Plos Biol., 22, 2024
|
|
4WZF
 
 | | Crystal structural basis for Rv0315, an immunostimulatory antigen and pseudo beta-1, 3-glucanase of Mycobacterium tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, 1,3-beta-glucanase, CALCIUM ION | | Authors: | Dong, W.Y, Fu, Z.F, Peng, G.Q. | | Deposit date: | 2014-11-19 | | Release date: | 2015-12-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Crystal structural basis for Rv0315, an immunostimulatory antigen and inactive beta-1,3-glucanase of Mycobacterium tuberculosis.
Sci Rep, 5, 2015
|
|
7KDT
 
 | |
7QG5
 
 | | IRAK4 in complex with inhibitor | | Descriptor: | 4-[(1-methylcyclopropyl)amino]-2-[[1-(1-methylpiperidin-4-yl)pyrazol-4-yl]amino]-6-pyrimidin-5-yl-pyrido[4,3-d]pyrimidin-5-one, Interleukin-1 receptor-associated kinase 4 | | Authors: | Xue, Y, Aagaard, A, Robb, G.R, Degorce, S.L. | | Deposit date: | 2021-12-07 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification and optimisation of a pyrimidopyridone series of IRAK4 inhibitors.
Bioorg.Med.Chem., 63, 2022
|
|
7QG2
 
 | | IRAK4 in complex with inhibitor | | Descriptor: | 6-methyl-4-[(1-methylcyclopropyl)amino]-2-[[1-(1-methylpiperidin-4-yl)pyrazol-4-yl]amino]pyrido[4,3-d]pyrimidin-5-one, Interleukin-1 receptor-associated kinase 4 | | Authors: | Xue, Y, Aagaard, A, Robb, G.R, Degorce, S.L. | | Deposit date: | 2021-12-07 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.031 Å) | | Cite: | Identification and optimisation of a pyrimidopyridone series of IRAK4 inhibitors.
Bioorg.Med.Chem., 63, 2022
|
|
6BOF
 
 | |
6CEV
 
 | | MBD3 MBD in complex with methylated, non-palindromic CpG DNA: alternative interpretation of crystallographic data | | Descriptor: | DNA (5'-D(*GP*CP*CP*AP*AP*(5CM)P*GP*CP*TP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*AP*GP*(5CM)P*GP*TP*TP*GP*GP*C)-3'), Methyl-CpG-binding domain protein 3, ... | | Authors: | Liu, K, Tempel, W, Wernimont, A.K, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-12 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Structural analyses reveal that MBD3 is a methylated CG binder.
Febs J., 286, 2019
|
|
6CEU
 
 | | MBD3 MBD in complex with methylated, non-palindromic CpG DNA: alternative interpretation of crystallographic data | | Descriptor: | DNA (5'-D(*GP*CP*CP*AP*AP*(5CM)P*GP*CP*TP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*AP*GP*(5CM)P*GP*TP*TP*GP*GP*C)-3'), Methyl-CpG-binding domain protein 3, ... | | Authors: | Liu, K, Tempel, W, Wernimont, A.K, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-12 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Structural analyses reveal that MBD3 is a methylated CG binder.
Febs J., 286, 2019
|
|
