2WSF
 
 | | Improved Model of Plant Photosystem I | | Descriptor: | 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, AT3G54890, BETA-CAROTENE, ... | | Authors: | Amunts, A, Toporik, H, Borovikov, A, Nelson, N. | | Deposit date: | 2009-09-05 | | Release date: | 2009-11-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.48 Å) | | Cite: | Structure determination and improved model of plant photosystem I.
J. Biol. Chem., 285, 2010
|
|
2KJZ
 
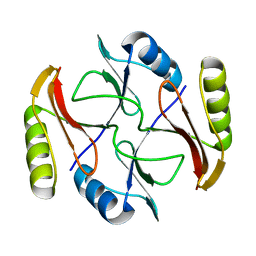 | | Solution NMR structure of protein ATC0852 from Agrobacterium tumefaciens. Northeast Structural Genomics Consortium (NESG) target AtT2. | | Descriptor: | ATC0852 | | Authors: | Lemak, A, Yee, A, Gutmanas, A, Fares, C, Garcia, M, Montelione, G, Arrowsmith, C, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-12 | | Release date: | 2009-07-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of protein ATC0852 from Agrobacterium tumefaciens
To be Published
|
|
6EUJ
 
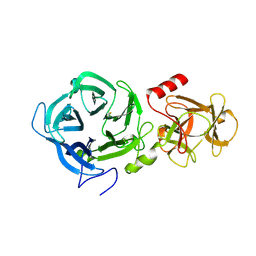 | | The GH43, Beta 1,3 Galactosidase, BT0265 | | Descriptor: | Beta-glucanase | | Authors: | Cartmell, A, Gilbert, H.J. | | Deposit date: | 2017-10-30 | | Release date: | 2018-10-17 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A surface endogalactanase in Bacteroides thetaiotaomicron confers keystone status for arabinogalactan degradation.
Nat Microbiol, 3, 2018
|
|
3ORH
 
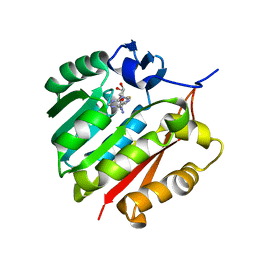 | | Human guanidinoacetate N-methyltransferase with SAH | | Descriptor: | Guanidinoacetate N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Dong, A, Wu, H, Zeng, H, Loppnau, P, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-09-07 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The crystal structure of human guanidinoacetate N-methyltransferase with SAH
TO BE PUBLISHED
|
|
3KVG
 
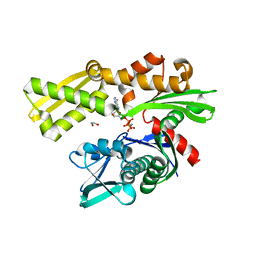 | | Crystal structure of the N-terminal domain of Hsp70 (cgd2_20) from Cryptosporidium parvum in complex with AMPPNP | | Descriptor: | 1,2-ETHANEDIOL, Heat shock 70 (HSP70) protein, PHOSPHATE ION, ... | | Authors: | Wernimont, A.K, Hutchinson, A, Sullivan, H, Lew, J, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Hui, R, Pizarro, J.C, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-11-30 | | Release date: | 2010-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of the N-terminal domain of Hsp70 (cgd2_20) from Cryptosporidium parvum in complex with AMPPNP
To be Published
|
|
7CCN
 
 | | The binding structure of a lanthanide binding tag (LBT3) with lutetium ion (Lu3+) | | Descriptor: | LBT3, LUTETIUM (III) ION | | Authors: | Hatanaka, T, Kikkawa, N, Matsugami, A, Hosokawa, Y, Hayashi, F, Ishida, N. | | Deposit date: | 2020-06-17 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The origins of binding specificity of a lanthanide ion binding peptide.
Sci Rep, 10, 2020
|
|
6SA8
 
 | | ring-like DARPin-Armadillo fusion H83_D01 | | Descriptor: | 1,2-ETHANEDIOL, LYS-ARG-LYS-ARG-LYS-ARG-LYS-ARG-LYS-ARG, ring-like DARPin-Armadillo fusion H83_D01 | | Authors: | Ernst, P, Honegger, A, van der Valk, F, Ewald, C, Mittl, P.R.E, Plucktun, A. | | Deposit date: | 2019-07-16 | | Release date: | 2019-11-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Rigid fusions of designed helical repeat binding proteins efficiently protect a binding surface from crystal contacts.
Sci Rep, 9, 2019
|
|
1Z6N
 
 | | 1.5 A Crystal Structure of a Protein of Unknown Function PA1234 from Pseudomonas aeruginosa | | Descriptor: | MAGNESIUM ION, hypothetical protein PA1234 | | Authors: | Zhang, R, Xu, L, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-03-22 | | Release date: | 2005-05-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.5A crystal structure of a hypothetical protein PA1234 from Pseudomonas aeruginosa
To be Published
|
|
2ERA
 
 | | RECOMBINANT ERABUTOXIN A, S8G MUTANT | | Descriptor: | ERABUTOXIN A | | Authors: | Gaucher, J.F, Menez, R, Arnoux, B, Menez, A, Ducruix, A. | | Deposit date: | 1997-06-25 | | Release date: | 1997-12-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | High resolution x-ray analysis of two mutants of a curaremimetic snake toxin
Eur.J.Biochem., 267, 2000
|
|
5EGR
 
 | | tRNA guanine transglycosylase (TGT) in complex with an Immucillin derivative | | Descriptor: | 1,2-ETHANEDIOL, 2-azanyl-7-[(2~{S},3~{R},5~{S})-5-(hydroxymethyl)-3-oxidanyl-pyrrolidin-2-yl]-3,5-dihydropyrrolo[3,2-d]pyrimidin-4-one, GLYCEROL, ... | | Authors: | Ehrmann, F.R, Heine, A, Klebe, G. | | Deposit date: | 2015-10-27 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Synthesis of an Immucillin Derivative as a Class of tRNA-Guanine Transglycosylase Inhibitors: Exploration of a Transition State Analogous Binding Mode
To be Published
|
|
6IV0
 
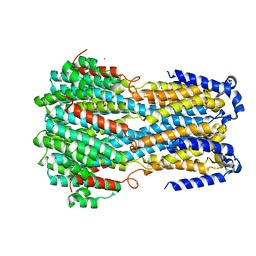 | |
6IVK
 
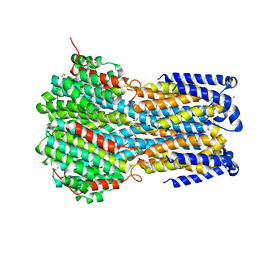 | | Crystal structure of a membrane protein G175A | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Kittredge, A, Fukuda, F, Zhang, Y, Yang, T. | | Deposit date: | 2018-12-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Dual Ca2+-dependent gates in human Bestrophin1 underlie disease-causing mechanisms of gain-of-function mutations.
Commun Biol, 2, 2019
|
|
6IVW
 
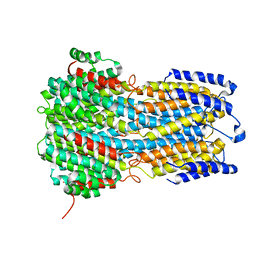 | |
3ZN1
 
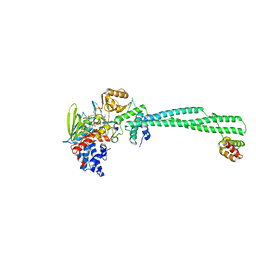 | | LSD1-CoREST in complex with PRLYLV peptide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, PEPTIDE, ... | | Authors: | Tortorici, M, Borrello, M.T, Tardugno, M, Chiarelli, L.R, Pilotto, S, Ciossani, G, Vellore, N.A, Cowan, J, O'Connell, M, Mai, A, Baron, R, Ganesan, A, Mattevi, A. | | Deposit date: | 2013-02-13 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Protein Recognition by Small Peptide Reversible Inhibitors of the Chromatin-Modifying Lsd1/Corest Lysine Demethylase.
Acs Chem.Biol., 8, 2013
|
|
3P1W
 
 | | Crystal Structure of RAB GDI from Plasmodium Falciparum, PFL2060c | | Descriptor: | RabGDI protein | | Authors: | Wernimont, A.K, Neculai, A.M, Weadge, J, MacKenzie, F, Cossar, D, Tempel, W, Bochkarev, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Langsley, G, Bosch, J, Hui, R, Pizzaro, J.C, Hutchinson, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-09-30 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of RAB GDI from Plasmodium Falciparum, PFL2060c
To be published
|
|
3P5W
 
 | | Actinidin from Actinidia arguta planch (Sarusashi) | | Descriptor: | Actinidin, CADMIUM ION | | Authors: | Manickam, Y, Nirmal, N, Suzuki, A, Sugiyama, Y, Yamane, T, Devadasan, V, Sharma, A. | | Deposit date: | 2010-10-11 | | Release date: | 2010-11-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural analysis of actinidin and a comparison of cadmium and sulfur anomalous signals from actinidin crystals measured using in-house copper- and chromium-anode X-ray sources
Acta Crystallogr.,Sect.D, 66, 2010
|
|
6IFG
 
 | | Crystal structure of M1 zinc metallopeptidase E323A mutant bound to Tyr-ser-ala substrate from Deinococcus radiodurans | | Descriptor: | FORMIC ACID, Tripeptides (TYR-SER-ALA), ZINC ION, ... | | Authors: | Agrawal, R, Kumar, A, Kumar, A, Makde, R.D. | | Deposit date: | 2018-09-20 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Two-domain aminopeptidase of M1 family: Structural features for substrate binding and gating in absence of C-terminal domain.
J.Struct.Biol., 208, 2019
|
|
6EUF
 
 | | The GH43, Beta 1,3 Galactosidase, BT0265 | | Descriptor: | Beta-glucanase, alpha-L-arabinofuranose-(1-3)-[alpha-L-arabinofuranose-(1-4)][beta-D-glucopyranuronic acid-(1-6)]beta-D-galactopyranose-(1-6)-beta-D-galactopyranose, alpha-L-rhamnopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-6)-[alpha-L-arabinofuranose-(1-3)][alpha-L-arabinofuranose-(1-4)]beta-D-galactopyranose-(1-6)-beta-D-galactopyranose | | Authors: | Cartmell, A, Gilbert, H.J. | | Deposit date: | 2017-10-30 | | Release date: | 2018-10-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A surface endogalactanase in Bacteroides thetaiotaomicron confers keystone status for arabinogalactan degradation.
Nat Microbiol, 3, 2018
|
|
4LQ6
 
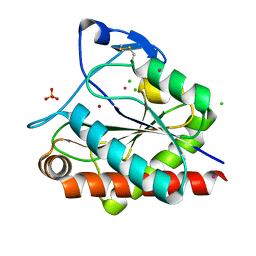 | | Crystal structure of Rv3717 reveals a novel amidase from M. tuberculosis | | Descriptor: | CHLORIDE ION, N-acetymuramyl-L-alanine amidase-related protein, PLATINUM (II) ION, ... | | Authors: | Kumar, A, Kumar, S, Kumar, D, Mishra, A, Dewangan, R.P, Shrivastava, P, Ramachandran, S, Taneja, B. | | Deposit date: | 2013-07-17 | | Release date: | 2013-12-04 | | Last modified: | 2014-01-15 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | The structure of Rv3717 reveals a novel amidase from Mycobacterium tuberculosis.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6EUH
 
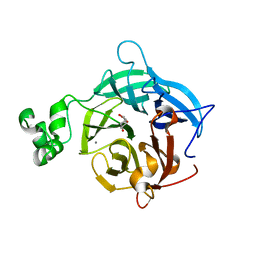 | | The GH43, Beta 1,3 Galactosidase, BT3683 with galactodeoxynojirimycin | | Descriptor: | (2R,3S,4R,5S)-2-(hydroxymethyl)piperidine-3,4,5-triol, Beta-glucanase, CALCIUM ION | | Authors: | Cartmell, A, Gilbert, H.J. | | Deposit date: | 2017-10-30 | | Release date: | 2018-10-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A surface endogalactanase in Bacteroides thetaiotaomicron confers keystone status for arabinogalactan degradation.
Nat Microbiol, 3, 2018
|
|
3KU9
 
 | |
3OUT
 
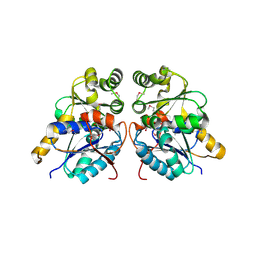 | | Crystal structure of glutamate racemase from Francisella tularensis subsp. tularensis SCHU S4 in complex with D-glutamate. | | Descriptor: | D-GLUTAMIC ACID, Glutamate racemase | | Authors: | Filippova, E.V, Wawrzak, Z, Onopriyenko, O, Kudriska, M, Edwards, A, Savchenko, A, Anderson, F.W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-15 | | Release date: | 2010-09-29 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of glutamate racemase from Francisella tularensis subsp. tularensis SCHU S4 in complex with D-glutamate.
To be Published
|
|
1W61
 
 | |
3ZF4
 
 | | Phage dUTPases control transfer of virulence genes by a proto- oncogenic G protein-like mechanism. (Staphylococcus bacteriophage 80alpha dUTPase Y81A mutant with dUpNHpp). | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, DUTPASE, MAGNESIUM ION, ... | | Authors: | Tormo-Mas, M.A, Donderis, J, Garcia-Caballer, M, Alt, A, Mir-Sanchis, I, Marina, A, Penades, J.R. | | Deposit date: | 2012-12-10 | | Release date: | 2013-01-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Phage Dutpases Control Transfer of Virulence Genes by a Proto-Oncogenic G Protein-Like Mechanism.
Mol.Cell, 49, 2013
|
|
3ZMV
 
 | | LSD1-CoREST in complex with PLSFLV peptide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, PKSFLV PEPTIDE, ... | | Authors: | Tortorici, M, Borrello, M.T, Tardugno, M, Chiarelli, L.R, Pilotto, S, Ciossani, G, Vellore, N.A, Cowan, J, O'Connell, M, Mai, A, Baron, R, Ganesan, A, Mattevi, A. | | Deposit date: | 2013-02-12 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Protein Recognition by Small Peptide Reversible Inhibitors of the Chromatin-Modifying Lsd1/Corest Lysine Demethylase.
Acs Chem.Biol., 8, 2013
|
|
