8EF9
 
 | | Structure of Lates calcarifer DNA polymerase theta polymerase domain with long duplex DNA, complex Ia | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*AP*GP*CP*AP*TP*CP*CP*GP*TP*AP*GP*(2DA))-3'), DNA (5'-D(*AP*GP*CP*TP*CP*TP*AP*CP*GP*GP*AP*TP*GP*C)-3'), ... | | Authors: | Li, C, Zhu, H, Sun, J, Gao, Y. | | Deposit date: | 2022-09-08 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural basis of DNA polymerase theta mediated DNA end joining.
Nucleic Acids Res., 51, 2023
|
|
8EFC
 
 | | Structure of Lates calcarifer DNA polymerase theta polymerase domain with long duplex DNA, complex Ia | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*AP*CP*TP*GP*TP*GP*AP*GP*GP*CP*AP*TP*CP*CP*GP*TP*AP*GP*(2DA))-3'), DNA (5'-D(*AP*GP*CP*TP*CP*TP*AP*CP*GP*GP*AP*TP*GP*CP*CP*TP*CP*AP*CP*AP*G)-3'), ... | | Authors: | Li, C, Zhu, H, Sun, J, Gao, Y. | | Deposit date: | 2022-09-08 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of DNA polymerase theta mediated DNA end joining.
Nucleic Acids Res., 51, 2023
|
|
8FH3
 
 | | Human IFT-A complex structures provide molecular insights into ciliary transport | | Descriptor: | Intraflagellar transport protein 122 homolog, Intraflagellar transport protein 140 homolog, Tubby-related protein 3, ... | | Authors: | Jiang, M, Palicharla, V.R, Miller, D, Hwang, S.H, Zhu, H, Hixson, P, Mukhopadhyay, S, Sun, J. | | Deposit date: | 2022-12-13 | | Release date: | 2023-02-22 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Human IFT-A complex structures provide molecular insights into ciliary transport.
Cell Res., 33, 2023
|
|
8FGW
 
 | | Human IFT-A complex structures provide molecular insights into ciliary transport | | Descriptor: | Intraflagellar transport protein 122 homolog, Intraflagellar transport protein 140 homolog, Intraflagellar transport protein 43 homolog, ... | | Authors: | Jiang, M, Palicharla, V.R, Miller, D, Hwang, S.H, Zhu, H, Hixson, P, Mukhopadhyay, S, Sun, J. | | Deposit date: | 2022-12-12 | | Release date: | 2023-02-22 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Human IFT-A complex structures provide molecular insights into ciliary transport.
Cell Res., 33, 2023
|
|
8IT0
 
 | | Cryo-EM structure of Crt-SPARTA-gRNA-tDNA dimer (conformation-2) | | Descriptor: | DNA (45-mer), Piwi domain-containing protein, RNA (5'-R(P*UP*GP*AP*GP*GP*UP*AP*GP*UP*AP*GP*GP*UP*UP*GP*UP*AP*UP*AP*GP*U)-3'), ... | | Authors: | Gao, X, Shang, K, Zhu, K, Wang, L, Mu, Z, Fu, X, Yu, X, Qin, B, Zhu, H, Ding, W, Cui, S. | | Deposit date: | 2023-03-21 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Nucleic-acid-triggered NADase activation of a short prokaryotic Argonaute.
Nature, 625, 2024
|
|
8ISZ
 
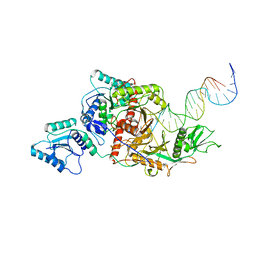 | | Cryo-EM structure of Crt-SPARTA-gRNA-tDNA monomer | | Descriptor: | DNA (45-mer), Piwi domain-containing protein, RNA (5'-R(P*UP*GP*AP*GP*GP*UP*AP*GP*UP*AP*GP*GP*UP*UP*GP*UP*AP*UP*AP*GP*U)-3'), ... | | Authors: | Gao, X, Shang, K, Zhu, K, Wang, L, Mu, Z, Fu, X, Yu, X, Qin, B, Zhu, H, Ding, W, Cui, S. | | Deposit date: | 2023-03-21 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Nucleic-acid-triggered NADase activation of a short prokaryotic Argonaute.
Nature, 625, 2024
|
|
8ISY
 
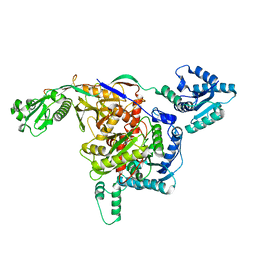 | | Cryo-EM structure of free-state Crt-SPARTA | | Descriptor: | Piwi domain-containing protein, TIR domain-containing protein | | Authors: | Gao, X, Shang, K, Zhu, K, Wang, L, Mu, Z, Fu, X, Yu, X, Qin, B, Zhu, H, Ding, W, Cui, S. | | Deposit date: | 2023-03-21 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Nucleic-acid-triggered NADase activation of a short prokaryotic Argonaute.
Nature, 625, 2024
|
|
8IT1
 
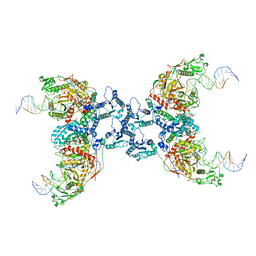 | | Cryo-EM structure of Crt-SPARTA-gRNA-tDNA tetramer (NADase active form) | | Descriptor: | DNA (45-mer), Piwi domain-containing protein, RNA (5'-R(P*UP*GP*AP*GP*GP*UP*AP*GP*UP*AP*GP*GP*UP*UP*GP*UP*AP*UP*AP*GP*U)-3'), ... | | Authors: | Gao, X, Shang, K, Zhu, K, Wang, L, Mu, Z, Fu, X, Yu, X, Qin, B, Zhu, H, Ding, W, Cui, S. | | Deposit date: | 2023-03-21 | | Release date: | 2023-11-08 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Nucleic-acid-triggered NADase activation of a short prokaryotic Argonaute.
Nature, 625, 2024
|
|
3V1S
 
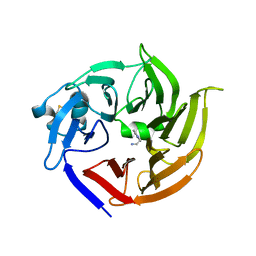 | | Scaffold tailoring by a newly detected Pictet-Spenglerase ac-tivity of strictosidine synthase (STR1): from the common tryp-toline skeleton to the rare piperazino-indole framework | | Descriptor: | 2-(1H-indol-1-yl)ethanamine, Strictosidine synthase | | Authors: | Wu, F, Zhu, H, Sun, L, Rajendran, C, Wang, M, Ren, X, Panjikar, S, Cherkasov, A, Zou, H, Stoeckigt, J. | | Deposit date: | 2011-12-10 | | Release date: | 2012-02-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.328 Å) | | Cite: | Scaffold Tailoring by a Newly Detected Pictet-Spenglerase Activity of Strictosidine Synthase: From the Common Tryptoline Skeleton to the Rare Piperazino-indole Framework
J.Am.Chem.Soc., 134, 2012
|
|
8K9G
 
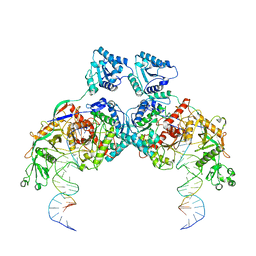 | | Cryo-EM structure of Crt-SPARTA-gRNA-tDNA dimer (conformation-1) | | Descriptor: | DNA (45-mer), Piwi domain-containing protein, RNA (5'-R(P*UP*GP*AP*GP*GP*UP*AP*GP*UP*AP*GP*GP*UP*UP*GP*UP*AP*UP*AP*GP*U)-3'), ... | | Authors: | Gao, X, Shang, K, Zhu, K, Wang, L, Mu, Z, Fu, X, Yu, X, Qin, B, Zhu, H, Ding, W, Cui, S. | | Deposit date: | 2023-08-01 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Nucleic-acid-triggered NADase activation of a short prokaryotic Argonaute.
Nature, 625, 2024
|
|
8X2Z
 
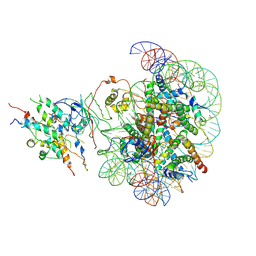 | | The class2 of piccolo NuA4 bound to the H2A.Z nucleosome complex at harboring state | | Descriptor: | Chromatin modification-related protein, DNA (146-MER), Histone H2A, ... | | Authors: | Wang, L, Zhang, H, Zhu, H, Zhu, P. | | Deposit date: | 2023-11-10 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures reveal the acetylation process of piccolo NuA4.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8X32
 
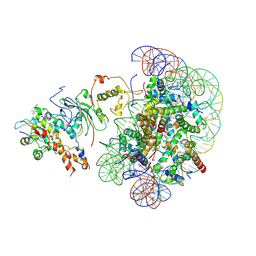 | | The piccolo NuA4 bound to the H2A.Z nucleosome-H4KQ Complex with Ac-CoA at resetting state | | Descriptor: | Chromatin modification-related protein, DNA (146-MER), Histone H2A, ... | | Authors: | Wang, L, Zhang, H, Zhu, H, Zhu, P. | | Deposit date: | 2023-11-10 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM structures reveal the acetylation process of piccolo NuA4.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8X30
 
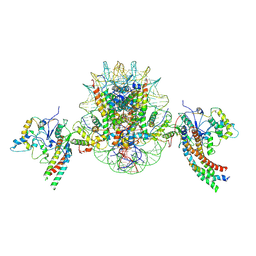 | | Structure of piccolo NuA4 and H2A.Z nucleosome 2:1 complex | | Descriptor: | Chromatin modification-related protein, DNA (146-MER), Histone H2A, ... | | Authors: | Wang, L, Zhang, H, Zhu, H, Zhu, P. | | Deposit date: | 2023-11-10 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM structures reveal the acetylation process of piccolo NuA4.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8X2X
 
 | | The piccolo NuA4 bound to the H2A.Z nucleosome complex at pre-H4-acetylation state | | Descriptor: | Chromatin modification-related protein, DNA (146-MER), Histone H2A, ... | | Authors: | Wang, L, Zhang, H, Zhu, H, Zhu, P. | | Deposit date: | 2023-11-10 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures reveal the acetylation process of piccolo NuA4.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8X2Y
 
 | | The class1 of piccolo NuA4 bound to the H2A.Z nucleosome complex at harboring state | | Descriptor: | Chromatin modification-related protein, DNA (146-MER), Histone H2A, ... | | Authors: | Wang, L, Zhang, H, Zhu, H, Zhu, P. | | Deposit date: | 2023-11-10 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structures reveal the acetylation process of piccolo NuA4.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8X31
 
 | | The piccolo NuA4 bound to the H2A.Z nucleosome complex with Ac-CoA at resetting state | | Descriptor: | Chromatin modification-related protein, DNA (146-MER), Histone H2A, ... | | Authors: | Wang, L, Zhang, H, Zhu, H, Zhu, P. | | Deposit date: | 2023-11-10 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Cryo-EM structures reveal the acetylation process of piccolo NuA4.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
6N5V
 
 | | Crystal Structure of Strictosidine in complex with 1H-indole-4-ethanamine | | Descriptor: | 2-(1H-indol-4-yl)ethan-1-amine, Strictosidine synthase | | Authors: | Cai, Y, Shao, N, Xie, H, Futamura, Y, Panjikar, S, Liu, H, Zhu, H, Osada, H, Zou, H. | | Deposit date: | 2018-11-22 | | Release date: | 2019-11-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.549 Å) | | Cite: | Crystal Structure of Strictosidine in complex with 1H-indole-4-ethanamine
to be published
|
|
6NJE
 
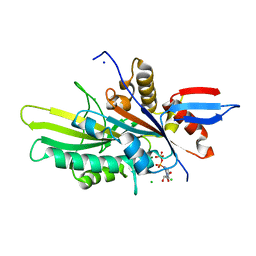 | | Crystal structure of the motor domain of human kinesin family member 22 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, Kinesin-like protein KIF22, ... | | Authors: | Walker, B.C, Zhu, H, Tempel, W, Arrowsmith, C.H, Edwards, A.M, Park, H, Cochran, J.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-01-03 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the motor domain of human kinesin family member 22
To Be Published
|
|
8ZDY
 
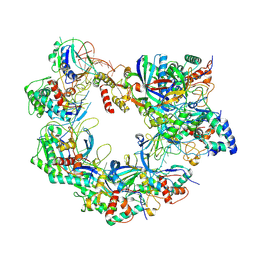 | | Cryo-EM structure of Cas8-HNH system at target free state | | Descriptor: | RNA (58-MER), a protein | | Authors: | Zhang, H, Zhu, H, Li, X, Liu, Y. | | Deposit date: | 2024-05-03 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for the type I-F Cas8-HNH system.
Embo J., 43, 2024
|
|
8Z0L
 
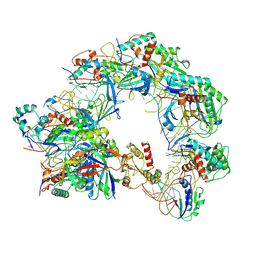 | | Cryo-EM structure of Cas8-HNH system at partial R-loop state | | Descriptor: | DNA (32-MER), DNA (5'-D(P*GP*TP*GP*CP*GP*GP*A)-3'), HNH endonuclease, ... | | Authors: | Zhang, H, Zhu, H, Li, X, Liu, Y. | | Deposit date: | 2024-04-09 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Structural basis for the type I-F Cas8-HNH system.
Embo J., 43, 2024
|
|
8Z0K
 
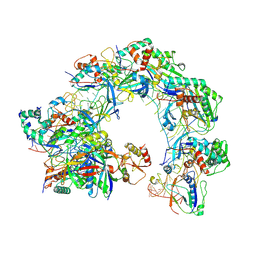 | | Cryo-EM structure of Cas8-HNH system at full R-loop state | | Descriptor: | DNA (37-MER), DNA (5'-D(P*GP*TP*GP*CP*GP*GP*A)-3'), HNH endonuclease, ... | | Authors: | Zhang, H, Zhu, H, Li, X, Liu, Y. | | Deposit date: | 2024-04-09 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Structural basis for the type I-F Cas8-HNH system.
Embo J., 43, 2024
|
|
8ZNR
 
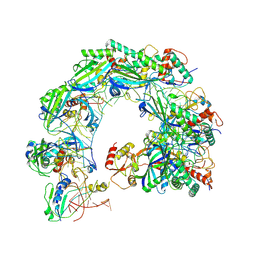 | | Cryo-EM structure of Cas8-HNH system at ssDNA-bound state | | Descriptor: | DNA (35-MER), RNA (56-MER), protein structure | | Authors: | Zhang, H, Zhu, H, Li, X, Liu, Y. | | Deposit date: | 2024-05-28 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for the type I-F Cas8-HNH system.
Embo J., 43, 2024
|
|
4IQ6
 
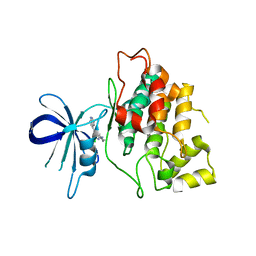 | | Gsk-3beta with inhibitor 6-chloro-N-cyclohexyl-4-(1H-pyrrolo[2,3-b]pyridin-3-yl)pyridin-2-amine | | Descriptor: | 6-chloro-N-cyclohexyl-4-(1H-pyrrolo[2,3-b]pyridin-3-yl)pyridin-2-amine, Glycogen synthase kinase-3 beta | | Authors: | Tong, Y, Stewart, K.D, Florjancic, A.S, Harlan, J.E, Merta, P.J, Przytulinska, M, Soni, N, Swinger, K.S, Zhu, H, Johnson, E.F, Shoemaker, A.R, Penning, T.D. | | Deposit date: | 2013-01-10 | | Release date: | 2013-04-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Azaindole-Based Inhibitors of Cdc7 Kinase: Impact of the Pre-DFG Residue, Val 195.
ACS Med Chem Lett, 4, 2013
|
|
8YQQ
 
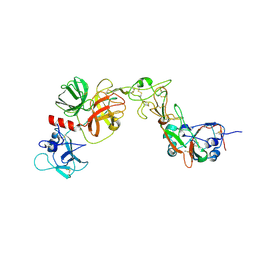 | | Structure of HKU1B RBD with TMPRSS2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, Transmembrane protease serine 2 | | Authors: | Gao, X, Cui, S, Ding, W, Zhu, K, Shang, K, Zhu, H. | | Deposit date: | 2024-03-19 | | Release date: | 2024-08-28 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Structural basis for the interaction between human coronavirus HKU1 spike receptor binding domain and its receptor TMPRSS2.
Cell Discov, 10, 2024
|
|
8YOY
 
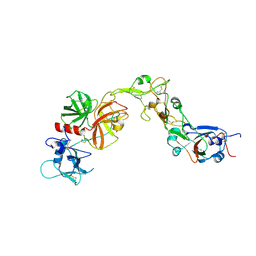 | | Structure of HKU1A RBD with TMPRSS2 | | Descriptor: | Spike protein S1, Transmembrane protease serine 2 | | Authors: | Gao, X, Cui, S, Ding, W, Shang, K, Zhu, H, Zhu, K. | | Deposit date: | 2024-03-14 | | Release date: | 2024-08-28 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structural basis for the interaction between human coronavirus HKU1 spike receptor binding domain and its receptor TMPRSS2.
Cell Discov, 10, 2024
|
|
