7ZK5
 
 | | ABCB1 L335C mutant (mABCB1) in the outward facing state bound to AAC | | Descriptor: | (4~{S},11~{S},18~{S})-4,11-dimethyl-18-(sulfanylmethyl)-6,13,20-trithia-3,10,17,22,23,24-hexazatetracyclo[17.2.1.1^{5,8}.1^{12,15}]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent translocase ABCB1, ... | | Authors: | Parey, K, Januliene, D, Gewering, T, Moeller, A. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Tracing the substrate translocation mechanism in P-glycoprotein.
Elife, 12, 2024
|
|
7ZK6
 
 | | ABCB1 L335C mutant (mABCB1) in the outward facing state bound to 2 molecules of AAC | | Descriptor: | (4S,11S,18S)-4-[[(2,4-dinitrophenyl)disulfanyl]methyl]-11,18-dimethyl-6,13,20-trithia-3,10,17,22,23,24-hexazatetracyclo[17.2.1.1^{5,8}.1^{12,15}]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, (4~{S},11~{S},18~{S})-4,11-dimethyl-18-(sulfanylmethyl)-6,13,20-trithia-3,10,17,22,23,24-hexazatetracyclo[17.2.1.1^{5,8}.1^{12,15}]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Parey, K, Januliene, D, Gewering, T, Moeller, A. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Tracing the substrate translocation mechanism in P-glycoprotein.
Elife, 12, 2024
|
|
7ZK9
 
 | | ABCB1 L971C mutant (mABCB1) in the inward facing state | | Descriptor: | (4~{S},11~{S},18~{S})-4,11-dimethyl-18-(sulfanylmethyl)-6,13,20-trithia-3,10,17,22,23,24-hexazatetracyclo[17.2.1.1^{5,8}.1^{12,15}]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, ATP-dependent translocase ABCB1 | | Authors: | Parey, K, Januliene, D, Gewering, T, Zhang, Q, Moeller, A. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Tracing the substrate translocation mechanism in P-glycoprotein.
Elife, 12, 2024
|
|
7ZK4
 
 | | The ABCB1 L335C mutant (mABCB1) in the outward facing state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent translocase ABCB1, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Parey, K, Januliene, D, Gewering, T, Moeller, A. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Tracing the substrate translocation mechanism in P-glycoprotein.
Elife, 12, 2024
|
|
7ZKB
 
 | | ABCB1 V978C mutant (mABCB1) in the inward facing state | | Descriptor: | (4S,11S,18S)-4-[[(2,4-dinitrophenyl)disulfanyl]methyl]-11,18-dimethyl-6,13,20-trithia-3,10,17,22,23,24-hexazatetracyclo[17.2.1.1^{5,8}.1^{12,15}]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, (4~{S},11~{S},18~{S})-4,11-dimethyl-18-(sulfanylmethyl)-6,13,20-trithia-3,10,17,22,23,24-hexazatetracyclo[17.2.1.1^{5,8}.1^{12,15}]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, ATP-dependent translocase ABCB1 | | Authors: | Parey, K, Januliene, D, Gewering, T, Moeller, A. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Tracing the substrate translocation mechanism in P-glycoprotein.
Elife, 12, 2024
|
|
7ZK8
 
 | | ABCB1 L971C mutant (mABCB1) in the outward facing state bound to AAC | | Descriptor: | (4~{S},11~{S},18~{S})-4,11-dimethyl-18-(sulfanylmethyl)-6,13,20-trithia-3,10,17,22,23,24-hexazatetracyclo[17.2.1.1^{5,8}.1^{12,15}]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent translocase ABCB1, ... | | Authors: | Parey, K, Januliene, D, Gewering, T, Moeller, A. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Tracing the substrate translocation mechanism in P-glycoprotein.
Elife, 12, 2024
|
|
8AVY
 
 | | The ABCB1 L335C mutant (mABCB1) in the Apo state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent translocase ABCB1, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Parey, K, Januliene, D, Gewering, T, Moeller, A. | | Deposit date: | 2022-08-27 | | Release date: | 2023-09-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Tracing the substrate translocation mechanism in P-glycoprotein.
Elife, 12, 2024
|
|
5GNK
 
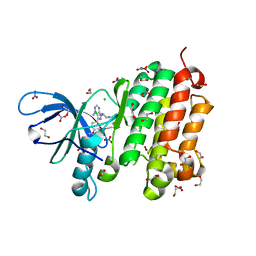 | | Crystal structure of EGFR 696-988 T790M in complex with LXX-6-34 | | Descriptor: | 1,2-ETHANEDIOL, 1-[(3R)-3-[4-azanyl-3-[3-chloranyl-4-[(1-methylimidazol-2-yl)methoxy]phenyl]pyrazolo[3,4-d]pyrimidin-1-yl]piperidin-1-yl]prop-2-en-1-one, CHLORIDE ION, ... | | Authors: | Yan, X.E, Yun, C.H. | | Deposit date: | 2016-07-21 | | Release date: | 2017-04-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.796 Å) | | Cite: | Discovery of (R)-1-(3-(4-Amino-3-(3-chloro-4-(pyridin-2-ylmethoxy)phenyl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl)piperidin-1-yl)prop-2-en-1-one (CHMFL-EGFR-202) as a Novel Irreversible EGFR Mutant Kinase Inhibitor with a Distinct Binding Mode.
J. Med. Chem., 60, 2017
|
|
5XDL
 
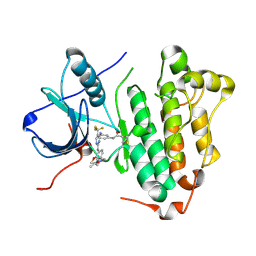 | | Crystal structure of EGFR 696-1022 L858R in complex with CO-1686 | | Descriptor: | Epidermal growth factor receptor, N-[3-[[2-[[4-(4-ethanoylpiperazin-1-yl)-2-methoxy-phenyl]amino]-5-(trifluoromethyl)pyrimidin-4-yl]amino]phenyl]prop-2-enamide | | Authors: | Yan, X.E, Zhu, S.J, Yun, C.H. | | Deposit date: | 2017-03-28 | | Release date: | 2017-12-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of mutant-selectivity and drug-resistance related to CO-1686.
Oncotarget, 8, 2017
|
|
8K3R
 
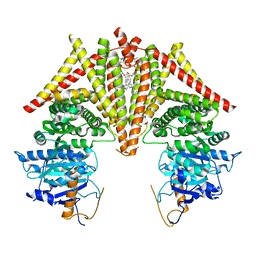 | | S. cerevisiae Chs1 in apo state incubated with GlcNAc | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Chitin synthase 1 | | Authors: | Bai, L, Chen, D. | | Deposit date: | 2023-07-16 | | Release date: | 2023-08-16 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Structure, catalysis, chitin transport, and selective inhibition of chitin synthase.
Nat Commun, 14, 2023
|
|
8K3U
 
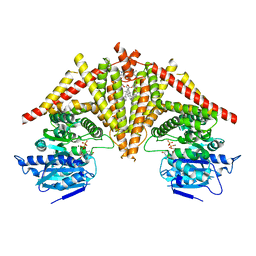 | | S. cerevisiae Chs1 in complex with UDP and GlcNAc | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, Chitin synthase 1, ... | | Authors: | Bai, L, Chen, D. | | Deposit date: | 2023-07-17 | | Release date: | 2023-09-06 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structure, catalysis, chitin transport, and selective inhibition of chitin synthase.
Nat Commun, 14, 2023
|
|
8K3W
 
 | | S. cerevisiae Chs1 in complex with UDP-GlcNAc and GlcNAc | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Chitin synthase 1, MAGNESIUM ION, ... | | Authors: | Bai, L, Chen, D. | | Deposit date: | 2023-07-17 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Structure, catalysis, chitin transport, and selective inhibition of chitin synthase.
Nat Commun, 14, 2023
|
|
8K3X
 
 | | S. cerevisiae Chs1 in complex with Nikkomycin Z | | Descriptor: | (2S)-{[(2S,3S,4S)-2-amino-4-hydroxy-4-(5-hydroxypyridin-2-yl)-3-methylbutanoyl]amino}[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxyoxolan-2-yl]acetic acid (non-preferred name), Chitin synthase 1 | | Authors: | Bai, L, Chen, D. | | Deposit date: | 2023-07-17 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structure, catalysis, chitin transport, and selective inhibition of chitin synthase.
Nat Commun, 14, 2023
|
|
8K3Q
 
 | | S. cerevisiae Chs1 in apo state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Chitin synthase 1 | | Authors: | Bai, L, Chen, D. | | Deposit date: | 2023-07-16 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure, catalysis, chitin transport, and selective inhibition of chitin synthase.
Nat Commun, 14, 2023
|
|
8K3V
 
 | | S. cerevisiae Chs1 in complex with UDP-GlcNAc | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Chitin synthase 1, MAGNESIUM ION, ... | | Authors: | Bai, L, Chen, D. | | Deposit date: | 2023-07-17 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure, catalysis, chitin transport, and selective inhibition of chitin synthase.
Nat Commun, 14, 2023
|
|
8K3T
 
 | | S. cerevisiae Chs1 in complex with UDP | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Chitin synthase 1, MAGNESIUM ION, ... | | Authors: | Bai, L, Chen, D. | | Deposit date: | 2023-07-17 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Structure, catalysis, chitin transport, and selective inhibition of chitin synthase.
Nat Commun, 14, 2023
|
|
8K3P
 
 | | S. cerevisiae Chs1 in complex with polyoxin B | | Descriptor: | (2S)-2-[[(2S,3S,4S)-5-aminocarbonyloxy-2-azanyl-3,4-bis(oxidanyl)pentanoyl]amino]-2-[(2R,3S,4R,5R)-5-[5-(hydroxymethyl)-2,4-bis(oxidanylidene)pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]ethanoic acid, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Chitin synthase 1 | | Authors: | Bai, L, Chen, D.D. | | Deposit date: | 2023-07-16 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structure, catalysis, chitin transport, and selective inhibition of chitin synthase.
Nat Commun, 14, 2023
|
|
8PEE
 
 | | ABCB1 L335C mutant (mABCB1) in the inward facing state bound to AAC | | Descriptor: | (4S,11S,18S)-4-[[(2,4-dinitrophenyl)disulfanyl]methyl]-11,18-dimethyl-6,13,20-trithia-3,10,17,22,23,24-hexazatetracyclo[17.2.1.1^{5,8}.1^{12,15}]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, (4~{S},11~{S},18~{S})-4,11-dimethyl-18-(sulfanylmethyl)-6,13,20-trithia-3,10,17,22,23,24-hexazatetracyclo[17.2.1.1^{5,8}.1^{12,15}]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, ATP-dependent translocase ABCB1, ... | | Authors: | Parey, K, Januliene, D, Gewering, T, Moeller, A. | | Deposit date: | 2023-06-13 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Tracing the substrate translocation mechanism in P-glycoprotein.
Elife, 12, 2024
|
|
7VEO
 
 | | Crystal structure of juvenile hormone acid methyltransferase silkworm JHAMT isoform3 complex with S-Adenosyl-L-homocysteine | | Descriptor: | Methyltranfer_dom domain-containing protein, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Guo, P.C, Zhang, Y.S, Zhang, l, Xu, H.Y. | | Deposit date: | 2021-09-09 | | Release date: | 2022-09-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structural characterization and functional analysis of juvenile hormone acid methyltransferase JHAMT3 from the silkworm, Bombyx mori.
Insect Biochem.Mol.Biol., 151, 2022
|
|
7XND
 
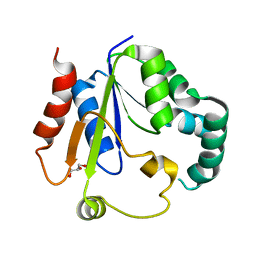 | | Crystal structure of Phosphomevalonate kinase from Silkworm | | Descriptor: | GLYCEROL, Phosphomevalonate kinase | | Authors: | Guo, P.C, Zhang, H. | | Deposit date: | 2022-04-28 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into the substrate binding of phosphomevalonate kinase from the silkworm, Bombyx mori.
Insect Biochem.Mol.Biol., 150, 2022
|
|
7V2S
 
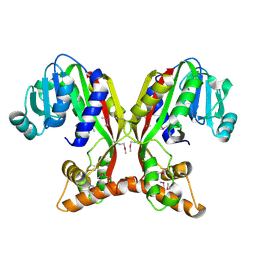 | | Crystal structure of juvenile hormone acid methyltransferase JHAMT isoform3 from silkworm | | Descriptor: | Methyltranfer_dom domain-containing protein | | Authors: | Guo, P.C, Zhang, Y.S, Zhang, l, Xu, H.Y. | | Deposit date: | 2021-08-09 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.133 Å) | | Cite: | Structural characterization and functional analysis of juvenile hormone acid methyltransferase JHAMT3 from the silkworm, Bombyx mori.
Insect Biochem.Mol.Biol., 151, 2022
|
|
7EBS
 
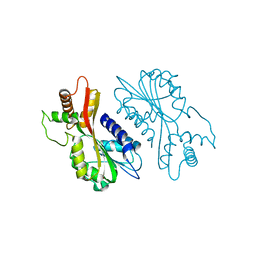 | | Crystal structure of juvenile hormone acid methyltransferase JHAMT from silkworm | | Descriptor: | Juvenile hormone acid methyltransferase | | Authors: | Guo, P.C, Zhang, Y.S, Zhang, L, Xu, H.Y, Xia, Q.Y. | | Deposit date: | 2021-03-11 | | Release date: | 2021-09-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural basis for juvenile hormone biosynthesis by the juvenile hormone acid methyltransferase.
J.Biol.Chem., 297, 2021
|
|
7EBX
 
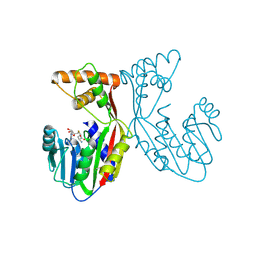 | | Crystal structure of juvenile hormone acid methyltransferase JHAMT in complex with S-adenosyl-L-homocysteine. | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, juvenile hormone acid methyltransferase | | Authors: | Guo, P.C, Zhang, Y.S, Zhang, L, Xu, H.Y, Xia, Q.Y. | | Deposit date: | 2021-03-11 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structural basis for juvenile hormone biosynthesis by the juvenile hormone acid methyltransferase.
J.Biol.Chem., 297, 2021
|
|
7EC0
 
 | | Crystal structure of juvenile hormone acid methyltransferase JHAMT in complex with S-Adenosyl homocysteine and methyl farnesoate | | Descriptor: | Juvenile hormone acid methyltransferase, Methyl farnesoate, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Guo, P.C, Zhang, Y.S, Zhang, L, Xu, H.Y, Xia, Q.Y. | | Deposit date: | 2021-03-11 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.494 Å) | | Cite: | Structural basis for juvenile hormone biosynthesis by the juvenile hormone acid methyltransferase.
J.Biol.Chem., 297, 2021
|
|
8HOU
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87A-T268V in complex with Im-N-C4-Phe-Phe | | Descriptor: | Bifunctional cytochrome P450/NADPH--P450 reductase, Im-N-C4-Phe-Phe, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2022-12-10 | | Release date: | 2023-12-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Anchoring a Structurally Editable Proximal Cofactor-like Module to Construct an Artificial Dual-center Peroxygenase.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
