6U0M
 
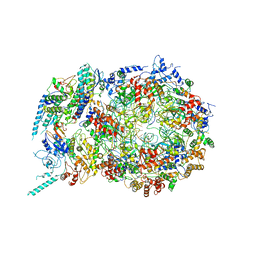 | | Structure of the S. cerevisiae replicative helicase CMG in complex with a forked DNA | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 45, DNA (15-MER), ... | | Authors: | Yuan, Z, Georgescu, R, Bai, L, Zhang, D, O'Donnell, M, Li, H. | | Deposit date: | 2019-08-14 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | DNA unwinding mechanism of a eukaryotic replicative CMG helicase.
Nat Commun, 11, 2020
|
|
2W0J
 
 | | Crystal structure of Chk2 in complex with NSC 109555, a specific inhibitor | | Descriptor: | 4,4'-DIACETYLDIPHENYLUREA-BIS(GUANYLHYDRAZONE), NITRATE ION, SERINE/THREONINE-PROTEIN KINASE CHK2 | | Authors: | Lountos, G.T, Tropea, J.E, Zhang, D, Jobson, A.G, Pommier, Y, Shoemaker, R.H, Waugh, D.S. | | Deposit date: | 2008-08-18 | | Release date: | 2009-02-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of Checkpoint Kinase 2 in Complex with Nsc 109555, a Potent and Selective Inhibitor
Protein Sci., 18, 2009
|
|
5ADY
 
 | | Cryo-EM structures of the 50S ribosome subunit bound with HflX | | Descriptor: | 23S RRNA, 50S RIBOSOMAL PROTEIN L1, 50S RIBOSOMAL PROTEIN L10, ... | | Authors: | Zhang, Y, Mandava, C.S, Cao, W, Li, X, Zhang, D, Li, N, Zhang, Y, Zhang, X, Qin, Y, Mi, K, Lei, J, Sanyal, S, Gao, N. | | Deposit date: | 2015-08-25 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Hflx is a Ribosome Splitting Factor Rescuing Stalled Ribosomes Under Stress Conditions
Nat.Struct.Mol.Biol., 22, 2015
|
|
1DTH
 
 | | METALLOPROTEASE | | Descriptor: | 4-(N-HYDROXYAMINO)-2R-ISOBUTYL-2S-(2-THIENYLTHIOMETHYL)SUCCINYL-L-PHENYLALANINE-N-METHYLAMIDE, ATROLYSIN C, CALCIUM ION, ... | | Authors: | Botos, I, Scapozza, L, Zhang, D, Liotta, L.A, Meyer, E.F. | | Deposit date: | 1996-02-12 | | Release date: | 1997-02-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Batimastat, a potent matrix mealloproteinase inhibitor, exhibits an unexpected mode of binding.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
4M3Q
 
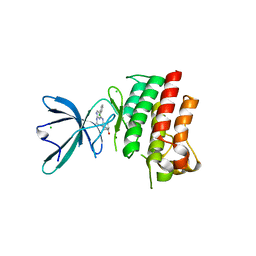 | | Crystal structure of the catalytic domain of the proto-oncogene tyrosine-protein kinase MER in complex with inhibitor UNC1917 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Tyrosine-protein kinase Mer, ... | | Authors: | Zhang, W, Zhang, D, Stashko, M.A, DeRyckere, D, Hunter, D, Kireev, D.B, Miley, M, Cummings, C, Lee, M, Norris-Drouin, J, Stewart, W.M, Sather, S, Zhou, Y, Kirkpatrick, G, Machius, M, Janzen, W.P, Earp, H.S, Graham, D.K, Frye, S, Wang, X. | | Deposit date: | 2013-08-06 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.718 Å) | | Cite: | Pseudo-Cyclization through Intramolecular Hydrogen Bond Enables Discovery of Pyridine Substituted Pyrimidines as New Mer Kinase Inhibitors.
J.Med.Chem., 56, 2013
|
|
6M1H
 
 | | CryoEM structure of human PAC1 receptor in complex with maxadilan | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Song, X, Wang, J, Zhang, D, Wang, H.W, Ma, Y. | | Deposit date: | 2020-02-26 | | Release date: | 2020-03-11 | | Last modified: | 2020-05-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures of PAC1 receptor reveal ligand binding mechanism.
Cell Res., 30, 2020
|
|
4PY0
 
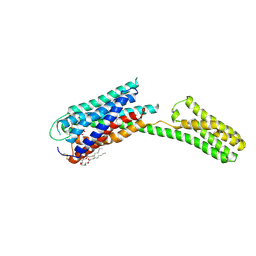 | | Crystal structure of P2Y12 receptor in complex with 2MeSATP | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-(methylsulfanyl)adenosine 5'-(tetrahydrogen triphosphate), P2Y purinoceptor 12, ... | | Authors: | Zhang, J, Zhang, K, Gao, Z.G, Paoletta, S, Zhang, D, Han, G.W, Li, T, Ma, L, Zhang, W, Muller, C.E, Yang, H, Jiang, H, Cherezov, V, Katritch, V, Jacobson, K.A, Stevens, R.C, Wu, B, Zhao, Q, GPCR Network (GPCR) | | Deposit date: | 2014-03-25 | | Release date: | 2014-04-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Agonist-bound structure of the human P2Y12 receptor
Nature, 509, 2014
|
|
4PXZ
 
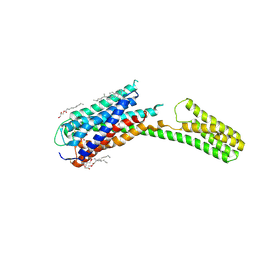 | | Crystal structure of P2Y12 receptor in complex with 2MeSADP | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-(methylsulfanyl)adenosine 5'-(trihydrogen diphosphate), CHOLESTEROL, ... | | Authors: | Zhang, J, Zhang, K, Gao, Z.G, Paoletta, S, Zhang, D, Han, G.W, Li, T, Ma, L, Zhang, W, Muller, C.E, Yang, H, Jiang, H, Cherezov, V, Katritch, V, Jacobson, K.A, Stevens, R.C, Wu, B, Zhao, Q, GPCR Network (GPCR) | | Deposit date: | 2014-03-25 | | Release date: | 2014-04-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Agonist-bound structure of the human P2Y12 receptor
Nature, 509, 2014
|
|
4NTJ
 
 | | Structure of the human P2Y12 receptor in complex with an antithrombotic drug | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHOLESTEROL, P2Y purinoceptor 12,Soluble cytochrome b562,P2Y purinoceptor 12, ... | | Authors: | Zhang, K, Zhang, J, Gao, Z.-G, Zhang, D, Zhu, L, Han, G.W, Moss, S.M, Paoletta, S, Kiselev, E, Lu, W, Fenalti, G, Zhang, W, Muller, C.E, Yang, H, Jiang, H, Cherezov, V, Katritch, V, Jacobson, K.A, Stevens, R.C, Wu, B, Zhao, Q, GPCR Network (GPCR) | | Deposit date: | 2013-12-02 | | Release date: | 2014-03-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structure of the human P2Y12 receptor in complex with an antithrombotic drug
Nature, 509, 2014
|
|
1U6D
 
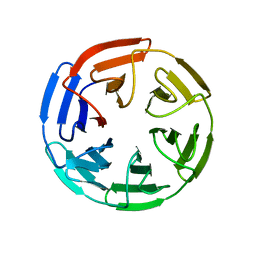 | |
3P2M
 
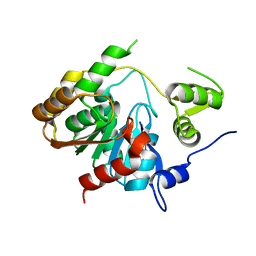 | | Crystal Structure of a Novel Esterase Rv0045c from Mycobacterium tuberculosis | | Descriptor: | POSSIBLE HYDROLASE | | Authors: | Zheng, X.D, Guo, J, Xu, L, Li, H, Zhang, D, Zhang, K, Sun, F, Wen, T, Liu, S, Pang, H. | | Deposit date: | 2010-10-03 | | Release date: | 2011-07-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of a Novel Esterase Rv0045c from Mycobacterium tuberculosis
Plos One, 6, 2011
|
|
4RIM
 
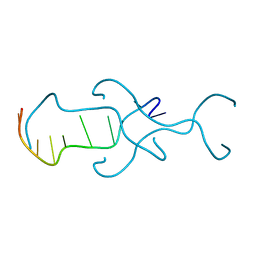 | |
4RIP
 
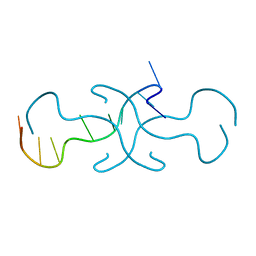 | |
1KS4
 
 | | The structure of Aspergillus niger endoglucanase-palladium complex | | Descriptor: | Endoglucanase A, PALLADIUM ION | | Authors: | Khademi, S, Zhang, D, Swanson, S.M, Wartenberg, A, Witte, C, Meyer, E.F. | | Deposit date: | 2002-01-10 | | Release date: | 2003-01-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Determination of the structure of an endoglucanase from Aspergillus niger and its mode of inhibition by palladium chloride.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1KS5
 
 | | Structure of Aspergillus niger endoglucanase | | Descriptor: | Endoglucanase A | | Authors: | Khademi, S, Zhang, D, Swanson, S.M, Wartenberg, A, Witte, C, Meyer, E.F. | | Deposit date: | 2002-01-10 | | Release date: | 2003-01-21 | | Last modified: | 2017-02-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Determination of the structure of an endoglucanase from Aspergillus niger and its mode of inhibition by palladium chloride.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
4FZ3
 
 | | Crystal structure of SIRT3 in complex with acetyl p53 peptide coupled with 4-amino-7-methylcoumarin | | Descriptor: | NAD-dependent protein deacetylase sirtuin-3, mitochondrial, ZINC ION, ... | | Authors: | Liu, D, Wu, J, Zhang, D, Chen, K, Jiang, H, Liu, H. | | Deposit date: | 2012-07-06 | | Release date: | 2013-03-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery and Mechanism Study of SIRT1 Activators that Promote the Deacetylation of Fluorophore-Labeled Substrate
J.Med.Chem., 56, 2013
|
|
8D1T
 
 | | Crystal structure of human USP30 in complex with a covalent inhibitor 552 and a Fab | | Descriptor: | (1R,2R,4S,7E)-7-[amino(sulfanyl)methylidene]-2-{[(1P)-3-chloro-3'-(1-cyanocyclopropyl)[1,1'-biphenyl]-4-carbonyl]amino}-7-azabicyclo[2.2.1]heptan-7-ium, 1,2-ETHANEDIOL, Ubiquitin carboxyl-terminal hydrolase 30, ... | | Authors: | Song, X, Butler, J, Li, C, Zhang, K, Zhang, D, Hao, Y. | | Deposit date: | 2022-05-27 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | TBD
To Be Published
|
|
6K7T
 
 | | Crystal structure of bat (Pteropus Alecto) MHC class I Ptal-N*01:01 in complex with Hendra virus-derived peptide HeV1--human beta-2 microglobulin | | Descriptor: | Beta-2-microglobulin, HeV1, MHC class I antigen | | Authors: | Lu, D, Liu, K.F, Zhang, D, Yue, C, Lu, Q, Cheng, H, Chai, Y, Qi, J.X, Gao, F.G, Liu, W.J. | | Deposit date: | 2019-06-08 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Peptide presentation by bat MHC class I provides new insight into the antiviral immunity of bats.
Plos Biol., 17, 2019
|
|
6K7U
 
 | | Crystal structure of beta-2 microglobulin (beta2m) of Bat (Pteropus Alecto) | | Descriptor: | Bat beta-2-microglobulin | | Authors: | Lu, D, Liu, K.F, Zhang, D, Yue, C, Lu, Q, Cheng, H, Chai, Y, Qi, J.X, Gao, F.G, Liu, W.J. | | Deposit date: | 2019-06-08 | | Release date: | 2019-09-18 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Peptide presentation by bat MHC class I provides new insight into the antiviral immunity of bats.
Plos Biol., 17, 2019
|
|
7QHB
 
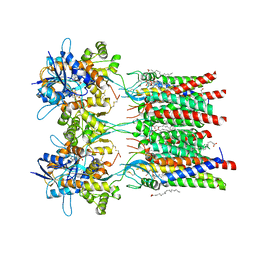 | | Active state of GluA1/2 in complex with TARP gamma 8, L-glutamate and CTZ | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (7Z)-hexadec-7-enoate, CYCLOTHIAZIDE, ... | | Authors: | Herguedas, B, Kohegyi, B, Zhang, D, Greger, I.H. | | Deposit date: | 2021-12-11 | | Release date: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mechanisms underlying TARP modulation of the GluA1/2-gamma 8 AMPA receptor.
Nat Commun, 13, 2022
|
|
7QHH
 
 | | Desensitized state of GluA1/2 AMPA receptor in complex with TARP-gamma 8 (TMD-LBD) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (7Z)-hexadec-7-enoate, GLUTAMIC ACID, ... | | Authors: | Herguedas, B, Kohegyi, B, Dohrke, J.N, Watson, J.F, Zhang, D, Ho, H, Shaikh, S, Lape, R, Krieger, J.M, Greger, I.H. | | Deposit date: | 2021-12-12 | | Release date: | 2022-02-23 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Mechanisms underlying TARP modulation of the GluA1/2-gamma 8 AMPA receptor.
Nat Commun, 13, 2022
|
|
6J23
 
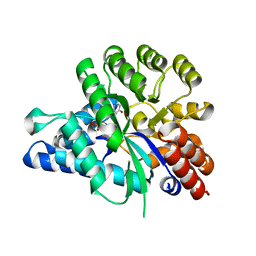 | | Crystal structure of arabidopsis ADAL complexed with GMP | | Descriptor: | Adenosine/AMP deaminase family protein, GUANOSINE-5'-MONOPHOSPHATE, ZINC ION | | Authors: | Wu, B.X, Zhang, D, Nie, H.B, Shen, S.L, Li, S.S, Patel, D.J. | | Deposit date: | 2018-12-30 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure ofArabidopsis thaliana N6-methyl-AMP deaminase ADAL with bound GMP and IMP and implications forN6-methyl-AMP recognition and processing.
Rna Biol., 16, 2019
|
|
6J4T
 
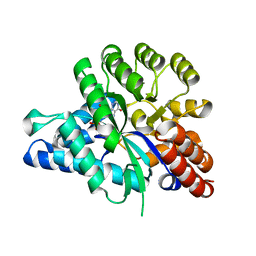 | | Crystal structure of arabidopsis ADAL complexed with IMP | | Descriptor: | Adenosine/AMP deaminase family protein, INOSINIC ACID, ZINC ION | | Authors: | Wu, B.X, Zhang, D, Nie, H.B, Shen, S.L, Li, S.S, Patel, D.J. | | Deposit date: | 2019-01-10 | | Release date: | 2019-07-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure ofArabidopsis thaliana N6-methyl-AMP deaminase ADAL with bound GMP and IMP and implications forN6-methyl-AMP recognition and processing.
Rna Biol., 16, 2019
|
|
6JRG
 
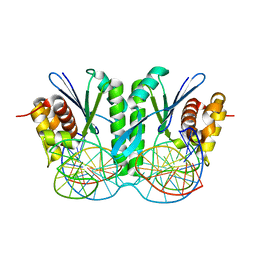 | | Crystal structure of ZmMoc1 H253A mutant in complex with Holliday junction | | Descriptor: | DNA (32-MER), DNA (33-MER), MAGNESIUM ION, ... | | Authors: | Lin, Z, Lin, H, Zhang, D, Yuan, C. | | Deposit date: | 2019-04-03 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Structural basis of sequence-specific Holliday junction cleavage by MOC1.
Nat.Chem.Biol., 15, 2019
|
|
6JRF
 
 | | Crystal structure of ZmMoc1-Holliday junction Complex in the presence of Calcium | | Descriptor: | CALCIUM ION, DNA (33-MER), Monokaryotic chloroplast 1, ... | | Authors: | Lin, Z, Lin, H, Zhang, D, Yuan, C. | | Deposit date: | 2019-04-03 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | Structural basis of sequence-specific Holliday junction cleavage by MOC1.
Nat.Chem.Biol., 15, 2019
|
|
