6C2T
 
 | | Aurora A ligand complex | | Descriptor: | (2S,4R)-1-[(3-chloro-2-fluorophenyl)methyl]-2-methyl-4-({3-[(1,3-thiazol-2-yl)amino]isoquinolin-1-yl}methyl)piperidine-4-carboxylic acid, Aurora kinase A, DIMETHYL SULFOXIDE, ... | | Authors: | Antonysamy, S, Pustilnik, A, Manglicmot, D, Froning, K, Weichert, K, Wasserman, S. | | Deposit date: | 2018-01-08 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Aurora A Kinase Inhibition Is Synthetic Lethal with Loss of theRB1Tumor Suppressor Gene.
Cancer Discov, 9, 2019
|
|
5ZVV
 
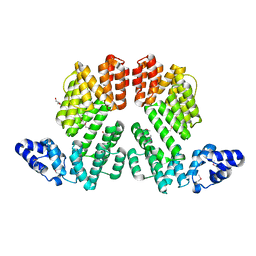 | | Structure of SeMet-phAimR | | Descriptor: | AimR transcriptional regulator, GLYCEROL | | Authors: | Cheng, W, Dou, C. | | Deposit date: | 2018-05-13 | | Release date: | 2018-09-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional insights into the regulation of the lysis-lysogeny decision in viral communities.
Nat Microbiol, 3, 2018
|
|
5ZW6
 
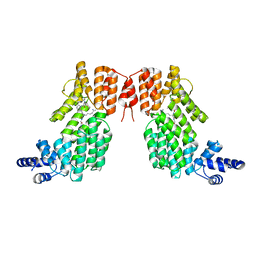 | | Structure of spAimR | | Descriptor: | AimR transcriptional regulator, GLY-MET-PRO-ARG-GLY-ALA | | Authors: | Cheng, W, Dou, C. | | Deposit date: | 2018-05-14 | | Release date: | 2018-09-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural and functional insights into the regulation of the lysis-lysogeny decision in viral communities.
Nat Microbiol, 3, 2018
|
|
5ZW5
 
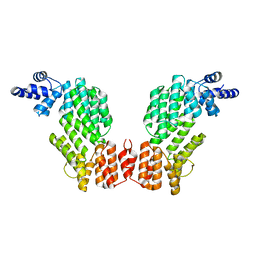 | | Structure of SeMet-spAimR | | Descriptor: | AimR transcriptional regulator | | Authors: | Cheng, W, Dou, C. | | Deposit date: | 2018-05-14 | | Release date: | 2018-08-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional insights into the regulation of the lysis-lysogeny decision in viral communities.
Nat Microbiol, 3, 2018
|
|
5ZVW
 
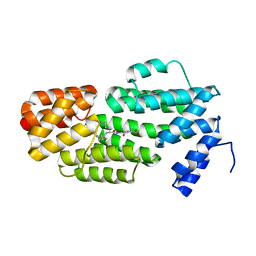 | | Structure of phAimR-Ligand | | Descriptor: | AimR transcriptional regulator, SER-ALA-ILE-ARG-GLY-ALA | | Authors: | Cheng, W, Dou, C. | | Deposit date: | 2018-05-13 | | Release date: | 2018-09-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.292 Å) | | Cite: | Structural and functional insights into the regulation of the lysis-lysogeny decision in viral communities.
Nat Microbiol, 3, 2018
|
|
2IBB
 
 | |
2H7T
 
 | |
2IBG
 
 | |
2IC2
 
 | |
7EDO
 
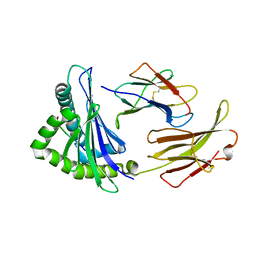 | | First insight into marsupial MHC I peptide presentation: immune features of lower mammals paralleled with bats | | Descriptor: | Beta-2-microglobulin, CYS-ASN-VAL-THR-LEU-ASN-TYR-PRO, MHC class I antigen | | Authors: | Wang, P.Y, Yue, C, Lu, D, Liu, K.F, Liu, S, Yao, S.J, Chai, Y, Qi, J.X, Lou, Y.L, Sun, Z.Y, Gao, G.F, Liu, W.J. | | Deposit date: | 2021-03-16 | | Release date: | 2021-08-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Peptide Presentations of Marsupial MHC Class I Visualize Immune Features of Lower Mammals Paralleled with Bats.
J Immunol., 207, 2021
|
|
8H2F
 
 | |
8HI1
 
 | | Streptococcus thermophilus Cas1-Cas2- prespacer ternary complex | | Descriptor: | CRISPR-associated endonuclease Cas1, DNA (26-MER), DNA (31-MER), ... | | Authors: | Chen, Q, Luo, Y. | | Deposit date: | 2022-11-18 | | Release date: | 2023-09-13 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | DnaQ mediates directional spacer acquisition in the CRISPR-Cas system by a time-dependent mechanism.
Innovation (N Y), 4, 2023
|
|
8H18
 
 | |
7CR8
 
 | | Synechocystis Cas1-Cas2-prespacerL complex | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2 1, DNA (36-MER) | | Authors: | Yu, Y, Chen, Q. | | Deposit date: | 2020-08-12 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Mechanisms of spacer acquisition by sequential assembly of the adaptation module in Synechocystis.
Nucleic Acids Res., 49, 2021
|
|
7CR6
 
 | | Synechocystis Cas1-Cas2/prespacer binary complex | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2 1, DNA (36-MER) | | Authors: | Yu, Y, Chen, Q. | | Deposit date: | 2020-08-12 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.72 Å) | | Cite: | Mechanisms of spacer acquisition by sequential assembly of the adaptation module in Synechocystis.
Nucleic Acids Res., 49, 2021
|
|
7XF3
 
 | | The structure of HLA-B*1501/BM58-66AF9 | | Descriptor: | 9-mer peptide from Matrix protein 1, Beta-2-microglobulin, MHC class I antigen | | Authors: | Zhao, Y.Z, Xiao, W.L, Wu, Y.N, Fan, W.F, Yue, C, Zhang, Q.X, Zhang, D.N, Yuan, X.J, Yao, S.J, Liu, S, Li, M, Wang, P.Y, Zhang, H.J, Zhang, J, Zhao, M, Zheng, X.Q, Liu, W.J, Gao, G.F, Liu, W.L. | | Deposit date: | 2022-03-31 | | Release date: | 2023-02-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Parallel T Cell Immunogenic Regions in Influenza B and A Viruses with Distinct Nuclear Export Signal Functions: The Balance between Viral Life Cycle and Immune Escape.
J Immunol., 210, 2023
|
|
7WKJ
 
 | | A COVID-19 T-cell response detection method based on a newly identified human CD8+ T cell epitope from SARS-CoV-2-Hubei Province, 2021. | | Descriptor: | Beta-2-microglobulin, LYS-THR-PHE-PRO-PRO-THR-GLU-PRO-LYS, MHC class I antigen | | Authors: | Zhang, J, Lu, D, Li, M, Liu, M.S, Yao, S.J, Zhan, J.B, Liu, J, Gao, G.F. | | Deposit date: | 2022-01-10 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A COVID-19 T-Cell Response Detection Method Based on a Newly Identified Human CD8 + T Cell Epitope from SARS-CoV-2 - Hubei Province, China, 2021.
China CDC Wkly, 4, 2022
|
|
3VNT
 
 | | Crystal Structure of the Kinase domain of Human VEGFR2 with a [1,3]thiazolo[5,4-b]pyridine derivative | | Descriptor: | 1,2-ETHANEDIOL, 2-chloro-3-(1-cyanocyclopropyl)-N-[5-({2-[(cyclopropylcarbonyl)amino][1,3]thiazolo[5,4-b]pyridin-5-yl}oxy)-2-fluorophenyl]benzamide, Vascular endothelial growth factor receptor 2 | | Authors: | Oki, H. | | Deposit date: | 2012-01-17 | | Release date: | 2012-04-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Design and synthesis of novel DFG-out RAF/vascular endothelial growth factor receptor 2 (VEGFR2) inhibitors. 1. Exploration of [5,6]-fused bicyclic scaffolds
J.Med.Chem., 55, 2012
|
|
4BDU
 
 | | Bax BH3-in-Groove dimer (GFP) | | Descriptor: | GREEN FLUORESCENT PROTEIN, APOPTOSIS REGULATOR BAX | | Authors: | Czabotar, P.E, Colman, P.M. | | Deposit date: | 2012-10-08 | | Release date: | 2013-02-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.998 Å) | | Cite: | Bax Crystal Structures Reveal How Bh3 Domains Activate Bax and Nucleate its Oligomerization to Induce Apoptosis.
Cell(Cambridge,Mass.), 152, 2013
|
|
4BD8
 
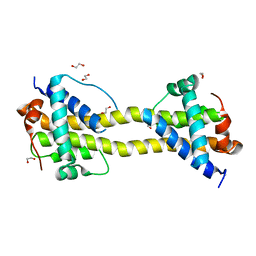 | | Bax domain swapped dimer induced by BimBH3 with CHAPS | | Descriptor: | 1,2-ETHANEDIOL, APOPTOSIS REGULATOR BAX, PRASEODYMIUM ION | | Authors: | Czabotar, P.E, Westphal, D, Adams, J.M, Colman, P.M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-02-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Bax Crystal Structures Reveal How Bh3 Domains Activate Bax and Nucleate its Oligomerization to Induce Apoptosis.
Cell(Cambridge,Mass.), 152, 2013
|
|
7EI1
 
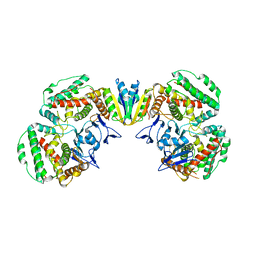 | | Structure of Pyrococcus furiosus Cas1Cas2 complex | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2 | | Authors: | Yu, Y, Chen, Q. | | Deposit date: | 2021-03-30 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | A distinct structure of Cas1-Cas2 complex provides insights into the mechanism for the longer spacer acquisition in Pyrococcus furiosus.
Int.J.Biol.Macromol., 183, 2021
|
|
7DPV
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with 7-O-methyl-dihydromyricetin | | Descriptor: | (2S,3S)-3,5-dihydroxy-7-methoxy-2-(3,4,5-trihydroxyphenyl)chroman-4-one, 3C-like proteinase | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Li, M.J, Xu, Y.C. | | Deposit date: | 2020-12-21 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Identification of pyrogallol as a warhead in design of covalent inhibitors for the SARS-CoV-2 3CL protease.
Nat Commun, 12, 2021
|
|
7DPP
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with myricetin | | Descriptor: | 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, 3C-like proteinase | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Li, M.J, Xu, Y.C. | | Deposit date: | 2020-12-21 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of pyrogallol as a warhead in design of covalent inhibitors for the SARS-CoV-2 3CL protease.
Nat Commun, 12, 2021
|
|
7DPU
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with 7-O-methyl-myricetin | | Descriptor: | 3C-like proteinase, 7-methoxy-3,5-bis(oxidanyl)-2-[3,4,5-tris(oxidanyl)phenyl]chromen-4-one, GLYCEROL | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Li, M.J, Xu, Y.C. | | Deposit date: | 2020-12-21 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Identification of pyrogallol as a warhead in design of covalent inhibitors for the SARS-CoV-2 3CL protease.
Nat Commun, 12, 2021
|
|
