7B18
 
 | | SARS-CoV-spike bound to two neutralising nanobodies | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody against SARS-CoV-2 VHH E, ... | | Authors: | Hallberg, B.M, Das, H. | | Deposit date: | 2020-11-24 | | Release date: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape.
Science, 371, 2021
|
|
6UUD
 
 | | Crystal structure of antibody 5D5 in complex with PfCSP N-terminal peptide | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 5D5 Antibody Fab, ... | | Authors: | Thai, E, Scally, S.W, Julien, J.P. | | Deposit date: | 2019-10-30 | | Release date: | 2020-07-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A high-affinity antibody against the CSP N-terminal domain lacks Plasmodium falciparum inhibitory activity.
J.Exp.Med., 217, 2020
|
|
6WCR
 
 | |
7KSG
 
 | | SARS-CoV-2 spike in complex with nanobodies E | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody against SARS-CoV-2 glycoprotein, Spike glycoprotein | | Authors: | Hallberg, B.M, Das, H. | | Deposit date: | 2020-11-22 | | Release date: | 2021-01-20 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape.
Science, 371, 2021
|
|
7KN3
 
 | | Crystal structure of SARS-CoV-2 spike protein receptor-binding domain complexed with a pre-pandemic antibody S-B8 Fab | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, S-B8 Fab heavy chain, ... | | Authors: | Liu, H, Zhu, X, Wilson, I.A. | | Deposit date: | 2020-11-04 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Neutralizing Antibodies to SARS-CoV-2 Selected from a Human Antibody Library Constructed Decades Ago.
Adv Sci, 9, 2022
|
|
7KN4
 
 | |
7LOP
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with antibodies CV05-163 and CR3022 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CR3022 Fab heavy chain, CR3022 Fab light chain, ... | | Authors: | Yuan, M, Zhu, X, Wilson, I.A. | | Deposit date: | 2021-02-10 | | Release date: | 2021-03-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.246 Å) | | Cite: | Structural and functional ramifications of antigenic drift in recent SARS-CoV-2 variants.
Science, 373, 2021
|
|
7LQ7
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with antibodies CV503 and COVA1-16 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COVA1-16 heavy chain, COVA1-16 light chain, ... | | Authors: | Yuan, M, Zhu, X, Wilson, I.A. | | Deposit date: | 2021-02-13 | | Release date: | 2021-09-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Bispecific antibodies targeting distinct regions of the spike protein potently neutralize SARS-CoV-2 variants of concern.
Sci Transl Med, 13, 2021
|
|
8FAQ
 
 | | Structure of Hemagglutinin from Influenza A/Victoria/22/2020 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Hernandez Garcia, A, Lei, R. | | Deposit date: | 2022-11-28 | | Release date: | 2024-06-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.035 Å) | | Cite: | Epistasis mediates the evolution of the receptor binding mode in recent human H3N2 hemagglutinin.
Nat Commun, 15, 2024
|
|
8FAW
 
 | |
6NC2
 
 | | AMC011 v4.2 SOSIP Env trimer in complex with fusion peptide targeting antibody ACS202 fragment antigen binding | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, AMC011 v4.2 SOSIP gp120, ... | | Authors: | Cottrell, C.A, Ozorowski, G, Yuan, M, Copps, J, Wilson, I.A, Ward, A.B. | | Deposit date: | 2018-12-10 | | Release date: | 2019-06-19 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Conformational Plasticity in the HIV-1 Fusion Peptide Facilitates Recognition by Broadly Neutralizing Antibodies.
Cell Host Microbe, 25, 2019
|
|
6NCP
 
 | | Crystal structure of HIV-1 broadly neutralizing antibody ACS202 | | Descriptor: | ACS202 Fab heavy chain, ACS202 Fab light chain, GLYCINE, ... | | Authors: | Yuan, M, Wilson, I.A. | | Deposit date: | 2018-12-11 | | Release date: | 2019-06-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Conformational Plasticity in the HIV-1 Fusion Peptide Facilitates Recognition by Broadly Neutralizing Antibodies.
Cell Host Microbe, 25, 2019
|
|
6NC3
 
 | | AMC011 v4.2 SOSIP Env trimer in complex with fusion peptide targeting antibody VRC34 fragment antigen binding | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 Env AMC011 v4.2 SOSIP gp120, ... | | Authors: | Cottrell, C.A, Ozorowski, G, Torres, J.L, Ward, A.B. | | Deposit date: | 2018-12-10 | | Release date: | 2019-06-19 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Conformational Plasticity in the HIV-1 Fusion Peptide Facilitates Recognition by Broadly Neutralizing Antibodies.
Cell Host Microbe, 25, 2019
|
|
5UXQ
 
 | | Structure of anti-HIV trimer apex antibody PGT143 | | Descriptor: | PGT143 Fab Heavy Chain, PGT143 Fab Light Chain | | Authors: | Kong, L, Wilson, I.A. | | Deposit date: | 2017-02-23 | | Release date: | 2017-04-19 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.415 Å) | | Cite: | A Broadly Neutralizing Antibody Targets the Dynamic HIV Envelope Trimer Apex via a Long, Rigidified, and Anionic Beta-Hairpin Structure
Immunity, 46, 2017
|
|
5UY3
 
 | | Crystal structure of human Fab PGT144, a broadly reactive and potent HIV-1 neutralizing antibody | | Descriptor: | Antibody PGT144 Fab heavy chain, Antibody PGT144 Fab light chain | | Authors: | Julien, J.-P, Lee, J.H, Wilson, I.A. | | Deposit date: | 2017-02-23 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A Broadly Neutralizing Antibody Targets the Dynamic HIV Envelope Trimer Apex via a Long, Rigidified, and Anionic beta-Hairpin Structure.
Immunity, 46, 2017
|
|
5V8M
 
 | | BG505 SOSIP.664 trimer in complex with broadly neutralizing HIV antibody 3BNC117 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lee, J.H, Cottrell, C.A, Ward, A.B. | | Deposit date: | 2017-03-22 | | Release date: | 2017-05-03 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | A Broadly Neutralizing Antibody Targets the Dynamic HIV Envelope Trimer Apex via a Long, Rigidified, and Anionic beta-Hairpin Structure.
Immunity, 46, 2017
|
|
6DCZ
 
 | |
6DCY
 
 | |
5V8L
 
 | | BG505 SOSIP.664 trimer in complex with broadly neutralizing HIV antibodies 3BNC117 and PGT145 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3BNC117 antibody, ... | | Authors: | Lee, J.H, Ward, A.B. | | Deposit date: | 2017-03-22 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | A Broadly Neutralizing Antibody Targets the Dynamic HIV Envelope Trimer Apex via a Long, Rigidified, and Anionic beta-Hairpin Structure.
Immunity, 46, 2017
|
|
6E0Q
 
 | | The N-terminal domain of PA endonuclease from the influenza H1N1 virus in complex with 4,6-dihydroxy-2-methyl-5-oxocyclohepta-1,3,6-triene-1-carboxylic acid | | Descriptor: | 1,2-ETHANEDIOL, 4,6-dihydroxy-2-methyl-5-oxocyclohepta-1,3,6-triene-1-carboxylic acid, MANGANESE (II) ION, ... | | Authors: | Dick, B.L, Morrison, C.N, Cohen, S.M. | | Deposit date: | 2018-07-06 | | Release date: | 2018-11-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure-Activity Relationships in Metal-Binding Pharmacophores for Influenza Endonuclease.
J. Med. Chem., 61, 2018
|
|
6DZQ
 
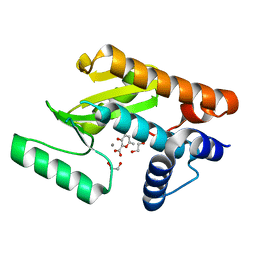 | | The N-terminal domain of PA endonuclease from the influenza H1N1 virus in complex with 3-hydroxy-6-methyl-4-oxo-4H-pyran-2-carboxylic acid | | Descriptor: | 1,2-ETHANEDIOL, 3-hydroxy-6-methyl-4-oxo-4H-pyran-2-carboxylic acid, MANGANESE (II) ION, ... | | Authors: | Dick, B.L, Morrison, C.N, Cohen, S.M. | | Deposit date: | 2018-07-05 | | Release date: | 2018-11-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-Activity Relationships in Metal-Binding Pharmacophores for Influenza Endonuclease.
J. Med. Chem., 61, 2018
|
|
7U4G
 
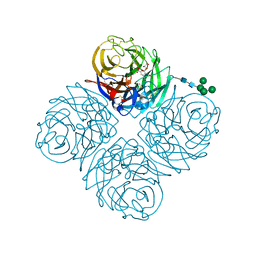 | | Neuraminidase from influenza virus A/Shandong/9/1993(H3N2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Neuraminidase, ... | | Authors: | Lei, R, Hernandez Garcia, A. | | Deposit date: | 2022-02-28 | | Release date: | 2022-10-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Prevalence and mechanisms of evolutionary contingency in human influenza H3N2 neuraminidase.
Nat Commun, 13, 2022
|
|
7U4F
 
 | |
7U4E
 
 | |
