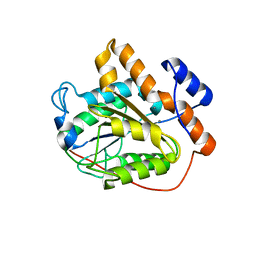
6K8A
 
 | |
7E4I
 
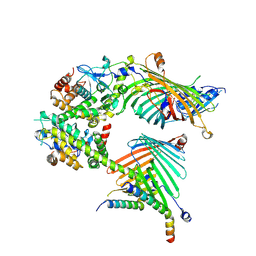 | | Cryo-EM structure of the yeast mitochondrial SAM-Tom40/Tom5/Tom6 complex at 3.0 angstrom | | Descriptor: | Mitochondrial import receptor subunit TOM40, Mitochondrial import receptor subunit TOM5, Mitochondrial import receptor subunit TOM6, ... | | Authors: | Wang, Q, Guan, Z.Y, Qi, L.B, Yan, C.Y, Yin, P. | | Deposit date: | 2021-02-13 | | Release date: | 2021-09-01 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural insight into the SAM-mediated assembly of the mitochondrial TOM core complex.
Science, 373, 2021
|
|
7E4H
 
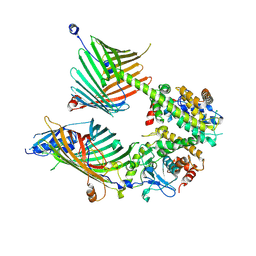 | | Cryo-EM structure of the yeast mitochondrial SAM-Tom40 complex at 3.0 angstrom | | Descriptor: | Mitochondrial import receptor subunit TOM40, Sorting assembly machinery 35 kDa subunit, Sorting assembly machinery 37 kDa subunit, ... | | Authors: | Wang, Q, Guan, Z.Y, Qi, L.B, Yan, C.Y, Yin, P. | | Deposit date: | 2021-02-13 | | Release date: | 2021-09-01 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural insight into the SAM-mediated assembly of the mitochondrial TOM core complex.
Science, 373, 2021
|
|
6K88
 
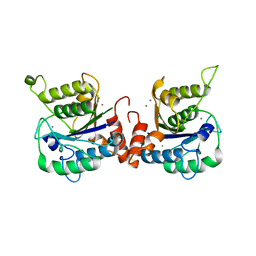 | | RGLG1 MIDAS binds calcium ion | | Descriptor: | CALCIUM ION, E3 ubiquitin-protein ligase RGLG1 | | Authors: | Wang, Q, Wu, Y. | | Deposit date: | 2019-06-11 | | Release date: | 2020-06-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.788 Å) | | Cite: | RGLG1 MIDAS binds calcium ion
To Be Published
|
|
8IHO
 
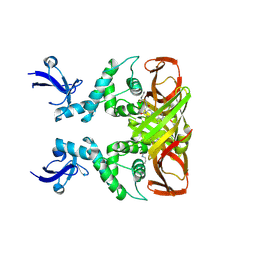 | | Crystal structures of SARS-CoV-2 papain-like protease in complex with covalent inhibitors | | Descriptor: | Papain-like protease nsp3, ZINC ION, covalent inhibitor | | Authors: | Wang, Q, Hu, H, Li, M, Xu, Y. | | Deposit date: | 2023-02-23 | | Release date: | 2024-01-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure-Based Design of Potent Peptidomimetic Inhibitors Covalently Targeting SARS-CoV-2 Papain-like Protease.
Int J Mol Sci, 24, 2023
|
|
8XT5
 
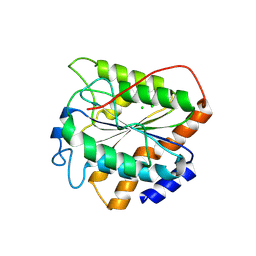 | | The closed state of RGLG5-VWA | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase RGLG5 | | Authors: | Wang, Q. | | Deposit date: | 2024-01-10 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.391 Å) | | Cite: | Structure of the E3 ubiquitin ligase RGLG5 VWA domain
To Be Published
|
|
7D58
 
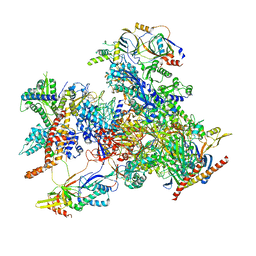 | | cryo-EM structure of human RNA polymerase III in elongating state | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Wang, Q, Wan, F, Lan, P, Wu, J, Lei, M. | | Deposit date: | 2020-09-25 | | Release date: | 2021-02-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into transcriptional regulation of human RNA polymerase III.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7D59
 
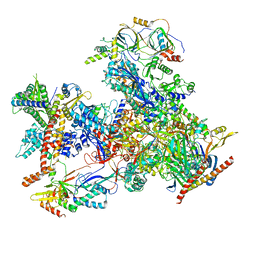 | | cryo-EM structure of human RNA polymerase III in apo state | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Wang, Q, Wan, F, Lan, P, Wu, J, Lei, M. | | Deposit date: | 2020-09-25 | | Release date: | 2021-02-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into transcriptional regulation of human RNA polymerase III.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7RPK
 
 | | Cryo-EM structure of murine Dispatched in complex with Sonic hedgehog | | Descriptor: | (2S)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(hexanoyloxy)propyl hexanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Asarnow, D, Wang, Q, Ding, K, Cheng, Y, Beachy, P.A. | | Deposit date: | 2021-08-03 | | Release date: | 2021-10-27 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Dispatched uses Na + flux to power release of lipid-modified Hedgehog.
Nature, 599, 2021
|
|
7RPJ
 
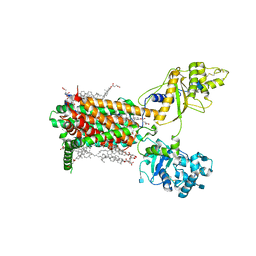 | | Cryo-EM structure of murine Dispatched NNN mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Protein dispatched homolog 1 | | Authors: | Asarnow, D, Wang, Q, Ding, K, Cheng, Y, Beachy, P.A. | | Deposit date: | 2021-08-03 | | Release date: | 2021-10-27 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Dispatched uses Na + flux to power release of lipid-modified Hedgehog.
Nature, 599, 2021
|
|
7RPH
 
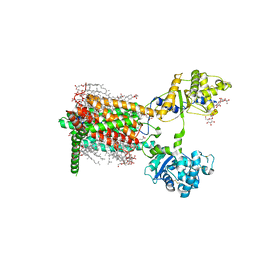 | | Cryo-EM structure of murine Dispatched 'R' conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Lauryl Maltose Neopentyl Glycol, ... | | Authors: | Asarnow, D, Wang, Q, Ding, K, Cheng, Y, Beachy, P.A. | | Deposit date: | 2021-08-03 | | Release date: | 2021-10-27 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Dispatched uses Na + flux to power release of lipid-modified Hedgehog.
Nature, 599, 2021
|
|
7RPI
 
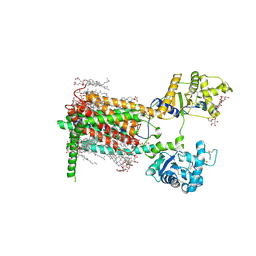 | | Cryo-EM structure of murine Dispatched 'T' conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Lauryl Maltose Neopentyl Glycol, ... | | Authors: | Asarnow, D, Wang, Q, Ding, K, Cheng, Y, Beachy, P.A. | | Deposit date: | 2021-08-03 | | Release date: | 2021-10-27 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Dispatched uses Na + flux to power release of lipid-modified Hedgehog.
Nature, 599, 2021
|
|
8HMH
 
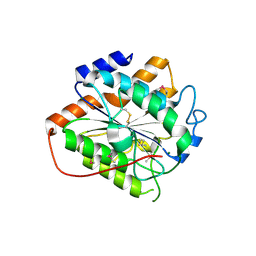 | | The closed state of RGLG2-VWA | | Descriptor: | E3 ubiquitin-protein ligase RGLG2, MAGNESIUM ION | | Authors: | Wang, Q. | | Deposit date: | 2022-12-03 | | Release date: | 2023-12-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | The regulation of RGLG2-VWA by Ca 2+ ions.
Biochim Biophys Acta Proteins Proteom, 1872, 2024
|
|
6VJT
 
 | |
3RWA
 
 | |
3RWT
 
 | |
8H3D
 
 | | Structure of apo SARS-CoV-2 spike protein with one RBD up | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin | | Authors: | Meng, F, Wang, Q, Xie, Y, Ni, X, Huang, N. | | Deposit date: | 2022-10-08 | | Release date: | 2023-03-22 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | In Silico Discovery of Small Molecule Modulators Targeting the Achilles' Heel of SARS-CoV-2 Spike Protein.
Acs Cent.Sci., 9, 2023
|
|
8H3E
 
 | | Complex structure of a small molecule (SPC-14) bound SARS-CoV-2 spike protein, closed state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 7-(6-nitro-2,3-dihydroindol-1-yl)-7-oxidanylidene-heptanoic acid, Spike glycoprotein,Fibritin | | Authors: | Meng, F, Wang, Q, Xie, Y, Ni, X, Huang, N. | | Deposit date: | 2022-10-08 | | Release date: | 2023-03-22 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | In Silico Discovery of Small Molecule Modulators Targeting the Achilles' Heel of SARS-CoV-2 Spike Protein.
Acs Cent.Sci., 9, 2023
|
|
7Y9J
 
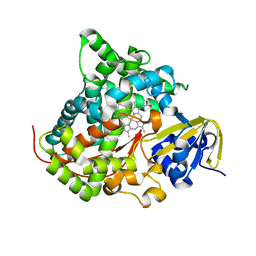 | | Crystal structure of P450 BM3-TMK from Bacillus megaterium in complex with 5-nitro-1,2-benzisoxazole | | Descriptor: | 5-nitro-1,2-benzoxazole, Bifunctional cytochrome P450/NADPH--P450 reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wang, Q, Zhang, L.L, Liu, W.D, Huang, J.-W, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-06-24 | | Release date: | 2023-06-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Engineering of a P450-based Kemp eliminase with a new mechanism
Chinese J Catal, 47, 2023
|
|
7Y9K
 
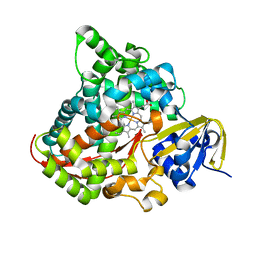 | | Crystal structure of P450 BM3-TMK from Bacillus megaterium | | Descriptor: | Bifunctional cytochrome P450/NADPH--P450 reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wang, Q, Zhang, L.L, Liu, W.D, Huang, J.-W, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-06-25 | | Release date: | 2023-06-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Engineering of a P450-based Kemp eliminase with a new mechanism
Chinese J Catal, 47, 2023
|
|
7DTE
 
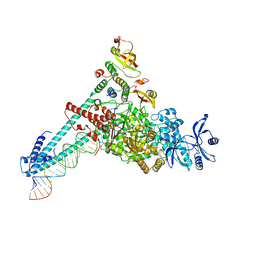 | | SARS-CoV-2 RdRP catalytic complex with T33-1 RNA | | Descriptor: | Non-structural protein 7, Non-structural protein 8, RNA (33-MER), ... | | Authors: | Wang, Q, Gong, P. | | Deposit date: | 2021-01-04 | | Release date: | 2021-10-20 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Remdesivir overcomes the S861 roadblock in SARS-CoV-2 polymerase elongation complex.
Cell Rep, 37, 2021
|
|
9IWV
 
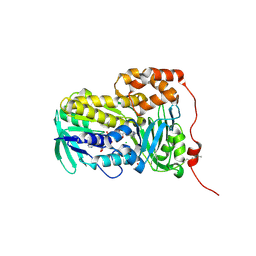 | | Crystal structure of Lsd18 after incubation with the substrate | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Wang, Q, Kim, C.Y, Chen, X. | | Deposit date: | 2024-07-26 | | Release date: | 2025-04-16 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mechanistic and molecular insights into iterative triepoxidation catalyzed by monooxygenase MonCI using a natural substrate analog.
Int.J.Biol.Macromol., 311, 2025
|
|
8YMK
 
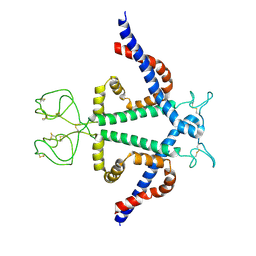 | | Localized reconstruction of Hepatitis B virus surface antigen dimer in the subviral particle with D2 symmetry from dataset A0 | | Descriptor: | Isoform S of Large envelope protein | | Authors: | Wang, T, Cao, L, Mu, A, Wang, Q, Rao, Z.H. | | Deposit date: | 2024-03-09 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Inherent symmetry and flexibility in hepatitis B virus subviral particles.
Science, 385, 2024
|
|
8YMJ
 
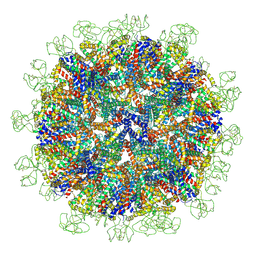 | | Cryo-EM structure of Hepatitis B virus surface antigen subviral particle with D2 symmetry | | Descriptor: | Isoform S of Large envelope protein | | Authors: | Wang, T, Cao, L, Mu, A, Wang, Q, Rao, Z.H. | | Deposit date: | 2024-03-09 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Inherent symmetry and flexibility in hepatitis B virus subviral particles.
Science, 385, 2024
|
|
4LLA
 
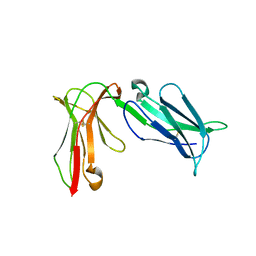 | | Crystal structure of D3D4 domain of the LILRB2 molecule | | Descriptor: | Leukocyte immunoglobulin-like receptor subfamily B member 2 | | Authors: | Nam, G, Shi, Y, Ryu, M, Wang, Q, Song, H, Liu, J, Yan, J, Qi, J, Gao, G.F. | | Deposit date: | 2013-07-09 | | Release date: | 2013-09-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Crystal structures of the two membrane-proximal Ig-like domains (D3D4) of LILRB1/B2: alternative models for their involvement in peptide-HLA binding
Protein Cell, 4, 2013
|
|
