3NTK
 
 | | Crystal structure of Tudor | | Descriptor: | Maternal protein tudor | | Authors: | Liu, H.P, Huang, Y, Li, Z.Z, Gong, W.M, Xu, R.M. | | Deposit date: | 2010-07-05 | | Release date: | 2010-09-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for methylarginine-dependent recognition of Aubergine by Tudor
Genes Dev., 24, 2010
|
|
4HTS
 
 | |
4XBJ
 
 | | Y274F alanine racemase from E. coli inhibited by l-ala-p | | Descriptor: | Alanine racemase, biosynthetic, SULFATE ION, ... | | Authors: | Squire, C.J, Yosaatmadja, Y, Patrick, W.M. | | Deposit date: | 2014-12-17 | | Release date: | 2015-12-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Mechanistic and Evolutionary Insights from the Reciprocal Promiscuity of Two Pyridoxal Phosphate-dependent Enzymes.
J.Biol.Chem., 291, 2016
|
|
3NTH
 
 | | Crystal structure of Tudor and Aubergine [R13(me2s)] complex | | Descriptor: | Maternal protein tudor, peptide from Aubergine | | Authors: | Liu, H.P, Huang, Y, Li, Z.Z, Gong, W.M, Xu, R.M. | | Deposit date: | 2010-07-05 | | Release date: | 2010-09-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for methylarginine-dependent recognition of Aubergine by Tudor
Genes Dev., 24, 2010
|
|
4HTT
 
 | |
3PRK
 
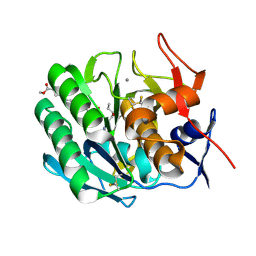 | | INHIBITION OF PROTEINASE K BY METHOXYSUCCINYL-ALA-ALA-PRO-ALA-CHLOROMETHYL KETONE. AN X-RAY STUDY AT 2.2-ANGSTROMS RESOLUTION | | Descriptor: | CALCIUM ION, METHOXYSUCCINYL-ALA-ALA-PRO-ALA-CHLOROMETHYL KETONE, PROTEINASE K | | Authors: | Wolf, W.M, Bajorath, J, Mueller, A, Raghunathan, S, Singh, T.P, Hinrichs, W, Saenger, W. | | Deposit date: | 1991-08-07 | | Release date: | 1994-01-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inhibition of proteinase K by methoxysuccinyl-Ala-Ala-Pro-Ala-chloromethyl ketone. An x-ray study at 2.2-A resolution.
J.Biol.Chem., 266, 1991
|
|
4YI7
 
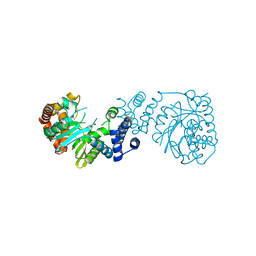 | |
3PKZ
 
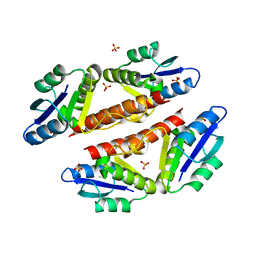 | | Structural basis for catalytic activation of a serine recombinase | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Recombinase Sin, ... | | Authors: | Keenholtz, R.A, Boocock, M.R, Rowland, S.J, Stark, W.M, Rice, P.A. | | Deposit date: | 2010-11-12 | | Release date: | 2011-06-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for catalytic activation of a serine recombinase.
Structure, 19, 2011
|
|
3QSB
 
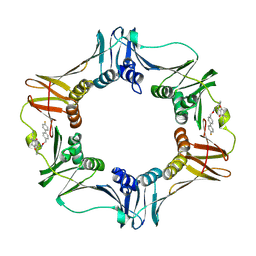 | | Structure of E. coli polIIIbeta with (Z)-5-(1-((4'-Fluorobiphenyl-4-yl)methoxyimino)butyl)-2,2-dimethyl-4,6-dioxocyclohexanecarbonitrile | | Descriptor: | (1R,5R)-5-{(1Z)-N-[(4'-fluorobiphenyl-4-yl)methoxy]butanimidoyl}-2,2-dimethyl-4,6-dioxocyclohexanecarbonitrile, DNA polymerase III subunit beta | | Authors: | Wijffels, G, Johnson, W.M, Oakley, A.J, Turner, K, Epa, V.C, Briscoe, S.J, Polley, M, Liepa, A.J, Hofmann, A, Buchardt, J, Christensen, C, Prosselkov, P, Dalrymple, B.P, Alewood, P.F, Jennings, P.A, Dixon, N.E, Winkler, D.A. | | Deposit date: | 2011-02-20 | | Release date: | 2011-06-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Binding inhibitors of the bacterial sliding clamp by design
J.Med.Chem., 54, 2011
|
|
3DMK
 
 | | Crystal structure of Down Syndrome Cell Adhesion Molecule (DSCAM) isoform 1.30.30, N-terminal eight Ig domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Down Syndrome Cell Adhesion Molecule (DSCAM) isoform 1.30.30, ... | | Authors: | Sawaya, M.R, Wojtowicz, W.M, Eisenberg, D, Zipursky, S.L. | | Deposit date: | 2008-07-01 | | Release date: | 2008-10-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (4.19 Å) | | Cite: | A double S shape provides the structural basis for the extraordinary binding specificity of Dscam isoforms.
Cell(Cambridge,Mass.), 134, 2008
|
|
8G01
 
 | | YES Complex - E. coli MraY, Protein E ID21, E. coli SlyD | | Descriptor: | FKBP-type peptidyl-prolyl cis-trans isomerase SlyD, GPE, Phospho-N-acetylmuramoyl-pentapeptide-transferase | | Authors: | Orta, A.K, Clemons, W.M, Riera, N. | | Deposit date: | 2023-01-31 | | Release date: | 2023-07-26 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The mechanism of the phage-encoded protein antibiotic from Phi X174.
Science, 381, 2023
|
|
8G02
 
 | | YES Complex - E. coli MraY, Protein E PhiX174, E. coli SlyD | | Descriptor: | Lysis protein E, Peptidyl-prolyl cis-trans isomerase, Phospho-N-acetylmuramoyl-pentapeptide-transferase | | Authors: | Orta, A.K, Clemons, W.M, Li, Y.E. | | Deposit date: | 2023-01-31 | | Release date: | 2023-07-26 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The mechanism of the phage-encoded protein antibiotic from Phi X174.
Science, 381, 2023
|
|
3ES5
 
 | | Crystal Structure of Partitivirus (PsV-F) | | Descriptor: | Putative capsid protein | | Authors: | Pan, J, Dong, L, Lin, L, Ochoa, W.F, Sinkovits, R.S, Havens, W.M, Nibert, M.L, Baker, T.S, Ghabrial, S.A, Tao, Y.J. | | Deposit date: | 2008-10-03 | | Release date: | 2009-03-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Atomic structure reveals the unique capsid organization of a dsRNA virus.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
6HJ5
 
 | | Crystal structure of Whitewater Arroyo virus GP1 glycoprotein at pH 5.6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Pre-glycoprotein polyprotein GP complex | | Authors: | Pryce, R, Ng, W.M, Zeltina, A, Watanabe, Y, El Omari, K, Wagner, A, Bowden, T.A. | | Deposit date: | 2018-08-31 | | Release date: | 2018-10-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structure-Based Classification Defines the Discrete Conformational Classes Adopted by the Arenaviral GP1.
J. Virol., 93, 2019
|
|
6HJC
 
 | | Crystal structure of Loei River virus GP1 glycoprotein at pH 8.0 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Pre-glycoprotein polyprotein GP complex | | Authors: | Pryce, R, Ng, W.M, Zeltina, A, Watanabe, Y, El Omari, K, Wagner, A, Bowden, T.A. | | Deposit date: | 2018-09-03 | | Release date: | 2018-10-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure-Based Classification Defines the Discrete Conformational Classes Adopted by the Arenaviral GP1.
J. Virol., 93, 2019
|
|
3F2R
 
 | | Crystal structure of human choline kinase alpha in complex with hemicholinium-3 | | Descriptor: | (2S,2'S)-2,2'-biphenyl-4,4'-diylbis(2-hydroxy-4,4-dimethylmorpholin-4-ium), Choline kinase alpha, UNKNOWN ATOM OR ION | | Authors: | Hong, B, Tempel, W, Rabeh, W.M, MacKenzie, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-10-30 | | Release date: | 2008-11-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of human choline kinase alpha in complex with hemicholinium-3
To be Published
|
|
6HJ6
 
 | | Crystal structure of Loei River virus GP1 glycoprotein at pH 5.0 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Pre-glycoprotein polyprotein GP complex | | Authors: | Pryce, R, Ng, W.M, Zeltina, A, Watanabe, Y, El Omari, K, Wagner, A, Bowden, T.A. | | Deposit date: | 2018-08-31 | | Release date: | 2018-10-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure-Based Classification Defines the Discrete Conformational Classes Adopted by the Arenaviral GP1.
J. Virol., 93, 2019
|
|
6JBS
 
 | | Bifunctional xylosidase/glucosidase LXYL | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gong, W.M, Yang, L.Y. | | Deposit date: | 2019-01-26 | | Release date: | 2020-02-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of beta-glycosidase LXYL-P1-2 reveals the product binding state of GH3 family and a specific pocket for Taxol recognition.
Commun Biol, 3, 2020
|
|
6KJ0
 
 | | Bifunctional xylosidase/glucosidase LXYL mutant E529Q C2221 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-D-xylosidase/beta-D-glucosidase, Deacetyltaxol, ... | | Authors: | Gong, W.M, Yang, L.Y. | | Deposit date: | 2019-07-20 | | Release date: | 2020-02-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structures of beta-glycosidase LXYL-P1-2 reveals the product binding state of GH3 family and a specific pocket for Taxol recognition.
Commun Biol, 3, 2020
|
|
3D7M
 
 | | Crystal Structure of the G Protein Fast-Exchange Double Mutant I56C/Q333C | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(i), alpha-1 subunit, ... | | Authors: | Funk, M.A, Preininger, A.M, Oldham, W.M, Meier, S.M, Hamm, H.E, Iverson, T.M. | | Deposit date: | 2008-05-21 | | Release date: | 2009-03-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Helix dipole movement and conformational variability contribute to allosteric GDP release in Galphai subunits.
Biochemistry, 48, 2009
|
|
5UXT
 
 | | Coiled-coil Trimer with Glu:Trp:Lys Triad | | Descriptor: | GLYCEROL, coiled-coil trimer with Glu:Trp:Lys triad | | Authors: | Smith, M.S, Billings, W.M, Whitby, F.G, Miller, M.B, Price, J.L. | | Deposit date: | 2017-02-23 | | Release date: | 2017-08-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | Enhancing a long-range salt bridge with intermediate aromatic and nonpolar amino acids.
Org. Biomol. Chem., 15, 2017
|
|
2FTL
 
 | | Crystal structure of trypsin complexed with BPTI at 100K | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cationic trypsin, ... | | Authors: | Hanson, W.M, Horvath, M.P, Goldenberg, D.P. | | Deposit date: | 2006-01-24 | | Release date: | 2006-02-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Rigidification of a Flexible Protease Inhibitor Variant upon Binding to Trypsin.
J.Mol.Biol., 366, 2007
|
|
2FV7
 
 | | Crystal structure of human ribokinase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ribokinase, ... | | Authors: | Rabeh, W.M, Tempel, W, Nedyalkova, L, Arrowsmith, C, Edwards, A, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-01-30 | | Release date: | 2006-02-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human ribokinase
to be published
|
|
2FTM
 
 | | Crystal structure of trypsin complexed with the BPTI variant (Tyr35->Gly) | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cationic trypsin, ... | | Authors: | Hanson, W.M, Horvath, M.P, Goldenberg, D.P. | | Deposit date: | 2006-01-24 | | Release date: | 2006-02-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Rigidification of a Flexible Protease Inhibitor Variant upon Binding to Trypsin.
J.Mol.Biol., 366, 2007
|
|
2H16
 
 | | Structure of human ADP-ribosylation factor-like 5 (ARL5) | | Descriptor: | ADP-ribosylation factor-like protein 5A, GUANOSINE-5'-DIPHOSPHATE, UNKNOWN ATOM OR ION | | Authors: | Rabeh, W.M, Tempel, W, Yaniw, D, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-05-16 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of human ADP-ribosylation factor-like 5 (ARL5)
To be Published
|
|
