6LUQ
 
 | | Haloperidol bound D2 dopamine receptor structure inspired discovery of subtype selective ligands | | Descriptor: | 4-[4-(4-chlorophenyl)-4-hydroxypiperidin-1-yl]-1-(4-fluorophenyl)butan-1-one, OLEIC ACID, chimera of D(2) dopamine receptor and Endolysin | | Authors: | Fan, L, Tan, L, Chen, Z, Qi, J, Nie, F, Luo, Z, Cheng, J, Wang, S. | | Deposit date: | 2020-01-30 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Haloperidol bound D2dopamine receptor structure inspired the discovery of subtype selective ligands.
Nat Commun, 11, 2020
|
|
4L6Y
 
 | | Structure of the microtubule associated protein PRC1 (Protein Regulator of Cytokinesis 1) | | Descriptor: | Protein regulator of cytokinesis 1 | | Authors: | Subramanian, R, Ti, S, Tan, L, Darst, S.A, Kapoor, T.M. | | Deposit date: | 2013-06-13 | | Release date: | 2013-07-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.3015 Å) | | Cite: | Marking and Measuring Single Microtubules by PRC1 and Kinesin-4.
Cell(Cambridge,Mass.), 154, 2013
|
|
4L3I
 
 | | Structure of the microtubule associated protein PRC1 (Protein Regulator of Cytokinesis 1) | | Descriptor: | Protein regulator of cytokinesis 1 | | Authors: | Subramanian, R, Ti, S, Tan, L, Darst, S.A, Kapoor, T.M. | | Deposit date: | 2013-06-06 | | Release date: | 2013-07-17 | | Last modified: | 2013-08-07 | | Method: | X-RAY DIFFRACTION (3.6005 Å) | | Cite: | Marking and Measuring Single Microtubules by PRC1 and Kinesin-4.
Cell(Cambridge,Mass.), 154, 2013
|
|
7F6J
 
 | | Crystal structure of the PDZD8 coiled-coil domain - Rab7 complex | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PDZ domain-containing protein 8, ... | | Authors: | Khan, H, Chen, L, Tan, L, Im, Y.J. | | Deposit date: | 2021-06-25 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of human PDZD8-Rab7 interaction for the ER-late endosome tethering.
Sci Rep, 11, 2021
|
|
7WWG
 
 | | Crystal structure of Saccharomyces cerevisiae Sfh2 complexed with phosphatidylinositol in an open conformation | | Descriptor: | (1R)-2-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}-1-[(octadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, Phosphatidylinositol transfer protein CSR1 | | Authors: | Chen, L, Tan, L, Im, Y.J. | | Deposit date: | 2022-02-12 | | Release date: | 2022-07-13 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural basis of ligand recognition and transport by Sfh2, a yeast phosphatidylinositol transfer protein of the Sec14 superfamily.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7WWD
 
 | | Crystal structure of Saccharomyces cerevisiae Sfh2 complexed with squalene | | Descriptor: | (6E,10E,14E,18E)-2,6,10,15,19,23-hexamethyltetracosa-2,6,10,14,18,22-hexaene, Phosphatidylinositol transfer protein CSR1 | | Authors: | Chen, L, Tan, L, Im, Y.J. | | Deposit date: | 2022-02-12 | | Release date: | 2022-07-13 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural basis of ligand recognition and transport by Sfh2, a yeast phosphatidylinositol transfer protein of the Sec14 superfamily.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7WWE
 
 | | Crystal structure of Saccharomyces cerevisiae Sfh2 in an apo form | | Descriptor: | Phosphatidylinositol transfer protein CSR1 | | Authors: | Chen, L, Tan, L, Im, Y.J. | | Deposit date: | 2022-02-12 | | Release date: | 2022-07-13 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of ligand recognition and transport by Sfh2, a yeast phosphatidylinositol transfer protein of the Sec14 superfamily.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7WVT
 
 | | Crystal structure of Saccharomyces cerevisiae Sfh2 complexed with phosphatidylinositol | | Descriptor: | (1R)-2-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}-1-[(octadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, Phosphatidylinositol transfer protein CSR1 | | Authors: | Chen, L, Tan, L, Im, Y.J. | | Deposit date: | 2022-02-11 | | Release date: | 2022-07-13 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of ligand recognition and transport by Sfh2, a yeast phosphatidylinositol transfer protein of the Sec14 superfamily.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7DEJ
 
 | | Structure of human ORP3 ORD in apo-form | | Descriptor: | Oxysterol-binding protein-related protein 3 | | Authors: | Tong, J, Tan, L, Im, Y.J. | | Deposit date: | 2020-11-04 | | Release date: | 2021-03-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of human ORP3 ORD reveals conservation of a key function and ligand specificity in OSBP-related proteins.
Plos One, 16, 2021
|
|
7DEI
 
 | | Structure of human ORP3 ORD domain in complex with PI(4)P | | Descriptor: | (2R)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5S,6R)-2,3,5,6-tetrahydroxy-4-(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dioctanoate, Oxysterol-binding protein-related protein 3 | | Authors: | Tong, J, Tan, L, Im, Y.J. | | Deposit date: | 2020-11-04 | | Release date: | 2021-03-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of human ORP3 ORD reveals conservation of a key function and ligand specificity in OSBP-related proteins.
Plos One, 16, 2021
|
|
5SBI
 
 | |
5SBJ
 
 | |
1OVU
 
 | |
3G9V
 
 | | Crystal structure of a soluble decoy receptor IL-22BP bound to interleukin-22 | | Descriptor: | Interleukin 22 receptor, alpha 2, Interleukin-22 | | Authors: | de Moura, P.R, Watanabe, L, Bleicher, L, Colau, D, Renauld, J.-C, Polikarpov, I. | | Deposit date: | 2009-02-14 | | Release date: | 2009-04-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.756 Å) | | Cite: | Crystal structure of a soluble decoy receptor IL-22BP bound to interleukin-22
FEBS Lett., 583, 2009
|
|
3DLQ
 
 | | Crystal structure of the IL-22/IL-22R1 complex | | Descriptor: | Interleukin-22, Interleukin-22 receptor subunit alpha-1 | | Authors: | Bleicher, L, de Moura, P.R, Watanabe, L, Colau, D, Dumoutier, L, Renauld, J.-C, Polikarpov, I. | | Deposit date: | 2008-06-28 | | Release date: | 2008-08-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the IL-22/IL-22R1 complex and its implications for the IL-22 signaling mechanism
Febs Lett., 582, 2008
|
|
3EOP
 
 | | Crystal Structure of the DUF55 domain of human thymocyte nuclear protein 1 | | Descriptor: | SULFATE ION, Thymocyte nuclear protein 1 | | Authors: | Yu, F, Song, A, Xu, C, Sun, L, Li, L, Tang, L, Hu, H, He, J. | | Deposit date: | 2008-09-29 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Determining the DUF55-domain structure of human thymocyte nuclear protein 1 from crystals partially twinned by tetartohedry
Acta Crystallogr.,Sect.D, 65, 2009
|
|
1OVV
 
 | |
3NB3
 
 | | The host outer membrane proteins OmpA and OmpC are packed at specific sites in the Shigella phage Sf6 virion as structural components | | Descriptor: | Outer membrane protein A, Outer membrane protein C | | Authors: | Zhao, H, Sequeira, R.D, Galeva, N.A, Tang, L. | | Deposit date: | 2010-06-02 | | Release date: | 2011-02-16 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (19 Å) | | Cite: | The host outer membrane proteins OmpA and OmpC are associated with the Shigella phage Sf6 virion.
Virology, 409, 2011
|
|
5L35
 
 | |
1OVR
 
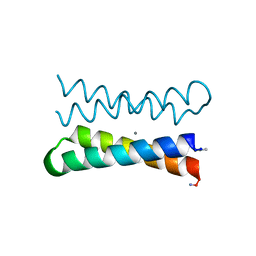 | | CRYSTAL STRUCTURE OF FOUR-HELIX BUNDLE MODEL di-Mn(II)-DF1-L13 | | Descriptor: | MANGANESE (II) ION, four-helix bundle model di-Mn(II)-DF1-L13 | | Authors: | Di Costanzo, L, Geremia, S. | | Deposit date: | 2003-03-27 | | Release date: | 2004-05-18 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Response of a designed metalloprotein to changes in metal ion coordination, exogenous ligands, and active site volume determined by X-ray crystallography.
J.Am.Chem.Soc., 127, 2005
|
|
3E6K
 
 | |
3KV2
 
 | |
6Q50
 
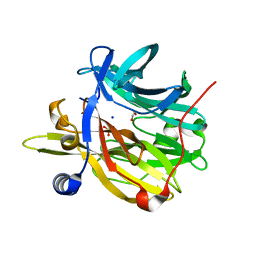 | | Structure of MPT-4, a mannose phosphorylase from Leishmania mexicana, in complex with phosphate ion | | Descriptor: | 1,2-ETHANEDIOL, MPT-4, PHOSPHATE ION, ... | | Authors: | Sobala, L.F, Males, A, Bastidas, L.M, Ward, T, Sernee, M.F, Ralton, J.E, Nero, T.L, Cobbold, S, Kloehn, J, Viera-Lara, M, Stanton, L, Hanssen, E, Parker, M.W, Williams, S.J, McConville, M.J, Davies, G.J. | | Deposit date: | 2018-12-06 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Family of Dual-Activity Glycosyltransferase-Phosphorylases Mediates Mannogen Turnover and Virulence in Leishmania Parasites.
Cell Host Microbe, 26, 2019
|
|
3LP7
 
 | |
2XQL
 
 | | Fitting of the H2A-H2B histones in the electron microscopy map of the complex Nucleoplasmin:H2A-H2B histones (1:5). | | Descriptor: | HISTONE H2A-IV, HISTONE H2B 5 | | Authors: | Ramos, I, Martin-Benito, J, Finn, R, Bretana, L, Aloria, K, Arizmendi, J.M, Ausio, J, Muga, A, Valpuesta, J.M, Prado, A. | | Deposit date: | 2010-09-02 | | Release date: | 2010-11-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (19.5 Å) | | Cite: | Nucleoplasmin Binds Histone H2A-H2B Dimers Through its Distal Face.
J.Biol.Chem., 285, 2010
|
|
