6WSB
 
 | |
6WSA
 
 | |
6N0Q
 
 | | BRAF in complex with N-(4-methyl-3-(1-methyl-2-oxo-2,3-dihydro-1H-benzo[d]imidazol-5-yl)phenyl)-3-(trifluoromethyl)benzamide. | | Descriptor: | N-[4-methyl-3-(1-methyl-2-oxo-2,3-dihydro-1H-benzimidazol-5-yl)phenyl]-3-(trifluoromethyl)benzamide, Serine/threonine-protein kinase B-raf | | Authors: | Mamo, M, Appleton, B.A. | | Deposit date: | 2018-11-07 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Design and Discovery ofN-(3-(2-(2-Hydroxyethoxy)-6-morpholinopyridin-4-yl)-4-methylphenyl)-2-(trifluoromethyl)isonicotinamide, a Selective, Efficacious, and Well-Tolerated RAF Inhibitor Targeting RAS Mutant Cancers: The Path to the Clinic.
J.Med.Chem., 63, 2020
|
|
6N0P
 
 | |
6WE5
 
 | |
5V0W
 
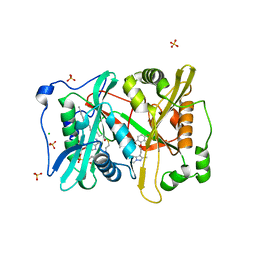 | | Crystal structure of glycylpeptide N-tetradecanoyltransferase from Plasmodium vivax in complex with inhibitor IMP-0001088 | | Descriptor: | 1-[5-[3,4-bis(fluoranyl)-2-[2-(1,3,5-trimethylpyrazol-4-yl)ethoxy]phenyl]-1-methyl-indazol-3-yl]-~{N},~{N}-dimethyl-methanamine, CHLORIDE ION, Glycylpeptide N-tetradecanoyltransferase, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2017-02-28 | | Release date: | 2018-03-07 | | Last modified: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Plasmodium vivax N-myristoyltransferase with inhibitor IMP-1088: exploring an NMT inhibitor for antimalarial therapy
Acta Crystallogr.,Sect.F, 81, 2025
|
|
6PQ3
 
 | | Crystal structure of GDP-bound KRAS with ten residues long internal tandem duplication in the switch II region | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Dharmaiah, S, Chan, A.H, Tran, T.H, Simanshu, D.K. | | Deposit date: | 2019-07-08 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | RASinternal tandem duplication disrupts GTPase-activating protein (GAP) binding to activate oncogenic signaling.
J.Biol.Chem., 295, 2020
|
|
7TMF
 
 | |
7TMG
 
 | |
8S6M
 
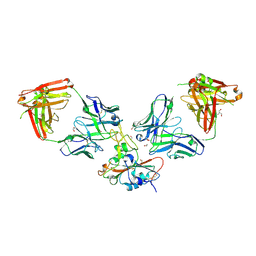 | | SARS-CoV-2 BQ.1.1 RBD bound to the S2V29 and the S2H97 Fab fragments | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Errico, J.M, Park, Y.J, Rietz, T, Czudnochowski, N, Nix, J.C, Cameroni, E, Corti, D, Snell, G, Marco, A.D, Pinto, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler, D. | | Deposit date: | 2024-02-28 | | Release date: | 2024-10-23 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | A potent pan-sarbecovirus neutralizing antibody resilient to epitope diversification.
Cell, 187, 2024
|
|
5UJ5
 
 | |
5VMT
 
 | |
6OK4
 
 | |
9D7K
 
 | | Infectious B19V capsid | | Descriptor: | Alpha-1-antichymotrypsin | | Authors: | Lee, H, Hafenstein, S. | | Deposit date: | 2024-08-16 | | Release date: | 2024-10-16 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Infectious parvovirus B19 circulates in the blood coated with active host protease inhibitors.
Nat Commun, 15, 2024
|
|
9C4N
 
 | | Infectious B19V capsid | | Descriptor: | Inter-alpha-trypsin inhibitor heavy chain H4 | | Authors: | Lee, H, Hafenstein, S. | | Deposit date: | 2024-06-04 | | Release date: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Infectious parvovirus B19 circulates in the blood coated with active host protease inhibitors.
Nat Commun, 15, 2024
|
|
9C27
 
 | | Infectious B19V capsid | | Descriptor: | Capsid protein 2 | | Authors: | Lee, H, Hafenstein, S. | | Deposit date: | 2024-05-30 | | Release date: | 2024-10-16 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Infectious parvovirus B19 circulates in the blood coated with active host protease inhibitors.
Nat Commun, 15, 2024
|
|
9C4F
 
 | | Infectious B19V capsid | | Descriptor: | Capsid protein 2, Inter-alpha-trypsin inhibitor heavy chain H4 | | Authors: | Lee, H, Hafenstein, S. | | Deposit date: | 2024-06-04 | | Release date: | 2024-10-16 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Infectious parvovirus B19 circulates in the blood coated with active host protease inhibitors.
Nat Commun, 15, 2024
|
|
9C2T
 
 | | Infectious B19V capsid | | Descriptor: | Alpha-1-antichymotrypsin His-Pro-less, Capsid protein 2 | | Authors: | Lee, H, Hafenstein, S. | | Deposit date: | 2024-05-31 | | Release date: | 2024-10-16 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Infectious parvovirus B19 circulates in the blood coated with active host protease inhibitors.
Nat Commun, 15, 2024
|
|
7KZF
 
 | |
3R20
 
 | |
6B8J
 
 | |
6B1Z
 
 | |
3K33
 
 | | Crystal structure of the Phd-Doc complex | | Descriptor: | ACETATE ION, Death on curing protein, Polypeptide of unknown amino acids and source, ... | | Authors: | Loris, R, Garcia-Pino, A. | | Deposit date: | 2009-10-01 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Allostery and intrinsic disorder mediate transcription regulation by conditional cooperativity.
Cell(Cambridge,Mass.), 142, 2010
|
|
7MU5
 
 | | Human DCTPP1 bound to Triptolide | | Descriptor: | MAGNESIUM ION, dCTP pyrophosphatase 1, triptolide | | Authors: | Hauk, G, Berger, J.M. | | Deposit date: | 2021-05-14 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Triptolide sensitizes cancer cells to nucleoside DNA methyltransferase inhibitors through inhibition of DCTPP1 mediated cell-intrinsic resistance
To Be Published
|
|
7NAF
 
 | | State E2 nucleolar 60S ribosomal biogenesis intermediate - Spb1-MTD local model | | Descriptor: | 25S rRNA, 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, ... | | Authors: | Cruz, V.E, Sekulski, K, Peddada, N, Erzberger, J.P. | | Deposit date: | 2021-06-21 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Sequence-specific remodeling of a topologically complex RNP substrate by Spb4.
Nat.Struct.Mol.Biol., 29, 2022
|
|
