4J31
 
 | | Crystal Structure of kynurenine 3-monooxygenase (KMO-396Prot) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Kynurenine 3-monooxygenase | | Authors: | Amaral, M, Levy, C, Heyes, D.J, Lafite, P, Outeiro, T.F, Giorgini, F, Leys, D, Scrutton, N.S. | | Deposit date: | 2013-02-05 | | Release date: | 2013-04-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of kynurenine 3-monooxygenase inhibition.
Nature, 496, 2013
|
|
4J36
 
 | | Cocrystal Structure of kynurenine 3-monooxygenase in complex with UPF 648 inhibitor(KMO-394UPF) | | Descriptor: | (1S,2S)-2-(3,4-dichlorobenzoyl)cyclopropanecarboxylic acid, FLAVIN-ADENINE DINUCLEOTIDE, Kynurenine 3-monooxygenase | | Authors: | Amaral, M, Levy, C, Heyes, D.J, Lafite, P, Outeiro, T.F, Giorgini, F, Leys, D, Scrutton, N.S. | | Deposit date: | 2013-02-05 | | Release date: | 2013-04-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural basis of kynurenine 3-monooxygenase inhibition.
Nature, 496, 2013
|
|
4J34
 
 | | Crystal Structure of kynurenine 3-monooxygenase - truncated at position 394 plus HIS tag cleaved. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Kynurenine 3-monooxygenase | | Authors: | Amaral, M, Levy, C, Heyes, D.J, Lafite, P, Outeiro, T.F, Giorgini, F, Leys, D, Scrutton, N.S. | | Deposit date: | 2013-02-05 | | Release date: | 2013-04-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural basis of kynurenine 3-monooxygenase inhibition.
Nature, 496, 2013
|
|
4J33
 
 | | Crystal Structure of kynurenine 3-monooxygenase (KMO-394) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Kynurenine 3-monooxygenase | | Authors: | Amaral, M, Levy, C, Heyes, D.J, Lafite, P, Outeiro, T.F, Giorgini, F, Leys, D, Scrutton, N.S. | | Deposit date: | 2013-02-05 | | Release date: | 2013-04-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural basis of kynurenine 3-monooxygenase inhibition.
Nature, 496, 2013
|
|
4KVR
 
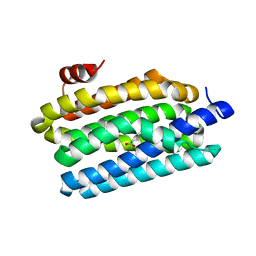 | | Crystal Structure of Prochlorococcus marinus aldehyde-deformylating oxygenase (mutant V41Y) | | Descriptor: | Aldehyde decarbonylase, FE (III) ION, HEXANOIC ACID | | Authors: | Levy, C.W, Khara, B, Menon, N, Mansell, D, Das, D, Marsh, E.N.G, Leys, D, Scrutton, N.S. | | Deposit date: | 2013-05-23 | | Release date: | 2013-06-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Production of propane and other short-chain alkanes by structure-based engineering of ligand specificity in aldehyde-deformylating oxygenase.
Chembiochem, 14, 2013
|
|
4KVS
 
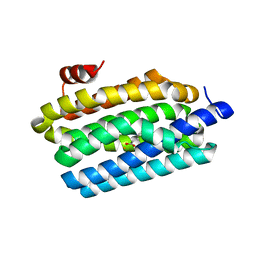 | | Crystal Structure of Prochlorococcus marinus aldehyde-deformylating oxygenase (mutant A134F) | | Descriptor: | Aldehyde decarbonylase, FE (III) ION, HEXANOIC ACID | | Authors: | Levy, C.W, Khara, B, Menon, N, Mansell, D, Das, D, Marsh, E.N.G, Leys, D, Scrutton, N.S. | | Deposit date: | 2013-05-23 | | Release date: | 2013-06-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Production of propane and other short-chain alkanes by structure-based engineering of ligand specificity in aldehyde-deformylating oxygenase.
Chembiochem, 14, 2013
|
|
4KVQ
 
 | | Crystal Structure of Prochlorococcus marinus aldehyde-deformylating oxygenase wild type with palmitic acid bound | | Descriptor: | Aldehyde decarbonylase, FE (III) ION, PALMITIC ACID | | Authors: | Levy, C.W, Khara, B, Menon, N, Mansell, D, Das, D, Marsh, E.N.G, Leys, D, Scrutton, N.S. | | Deposit date: | 2013-05-23 | | Release date: | 2013-06-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.842 Å) | | Cite: | Production of propane and other short-chain alkanes by structure-based engineering of ligand specificity in aldehyde-deformylating oxygenase.
Chembiochem, 14, 2013
|
|
5L53
 
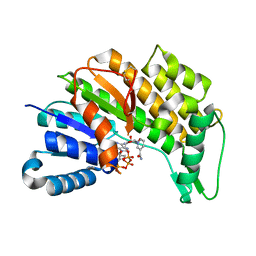 | | Menthone neomenthol reductase from Mentha piperita in complex with NADP | | Descriptor: | (-)-menthone:(+)-neomenthol reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Karuppiah, V, Toogood, H.S, Leys, D, Scrutton, N.S. | | Deposit date: | 2016-05-27 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Pinpointing a Mechanistic Switch Between Ketoreduction and "Ene" Reduction in Short-Chain Dehydrogenases/Reductases.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5L4S
 
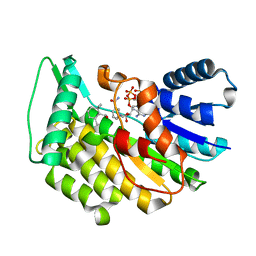 | | Isopiperitenone reductase from Mentha piperita in complex with NADP and beta-Cyclocitral | | Descriptor: | (-)-isopiperitenone reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, beta-cyclocitral | | Authors: | Karuppiah, V, Toogood, H.S, Leys, D, Scrutton, N.S. | | Deposit date: | 2016-05-26 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Pinpointing a Mechanistic Switch Between Ketoreduction and "Ene" Reduction in Short-Chain Dehydrogenases/Reductases.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5L51
 
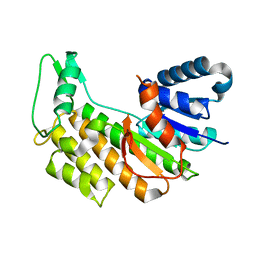 | |
5LDG
 
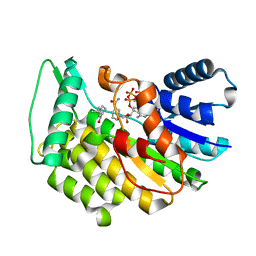 | | Isopiperitenone reductase from Mentha piperita in complex with Isopiperitenone and NADP | | Descriptor: | (-)-Isopiperitenone, (-)-isopiperitenone reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Karuppiah, V, Toogood, H.S, Leys, D, Scrutton, N.S. | | Deposit date: | 2016-06-26 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Pinpointing a Mechanistic Switch Between Ketoreduction and "Ene" Reduction in Short-Chain Dehydrogenases/Reductases.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5LCX
 
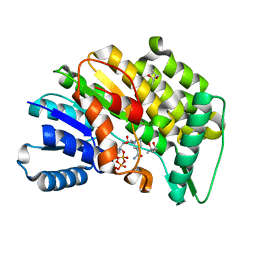 | | Isopiperitenone reductase from Mentha piperita in complex with NADP | | Descriptor: | (-)-isopiperitenone reductase, (4S)-2-METHYL-2,4-PENTANEDIOL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Karuppiah, V, Toogood, H.S, Leys, D, Scrutton, N.S. | | Deposit date: | 2016-06-22 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.709 Å) | | Cite: | Pinpointing a Mechanistic Switch Between Ketoreduction and "Ene" Reduction in Short-Chain Dehydrogenases/Reductases.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5N6Q
 
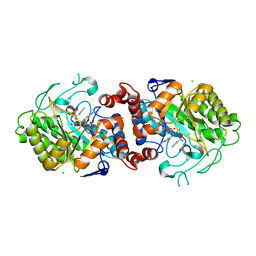 | | Xenobiotic reductase A (XenA) from Pseudomonas putida in complex with 2-phenylacrylic acid | | Descriptor: | 2-Phenylacrylic acid, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Karuppiah, V, Toogood, H.S, Leys, D, Scrutton, N.S. | | Deposit date: | 2017-02-15 | | Release date: | 2017-05-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into the ene-reductase synthesis of profens.
Org. Biomol. Chem., 15, 2017
|
|
5N6G
 
 | | NerA from Agrobacterium radiobacter in complex with 2-phenylacrylic acid | | Descriptor: | 2-Phenylacrylic acid, ACETATE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Karuppiah, V, Toogood, H.S, Leys, D, Scrutton, N.S. | | Deposit date: | 2017-02-15 | | Release date: | 2017-05-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural insights into the ene-reductase synthesis of profens.
Org. Biomol. Chem., 15, 2017
|
|
5NX4
 
 | |
5NX7
 
 | | Crystal structure of 1,8-cineole synthase from Streptomyces clavuligerus in complex with 2-fluoroneryl diphosphate and 2-fluorogeranyl diphosphate | | Descriptor: | (2E)-2-fluoro-3,7-dimethylocta-2,6-dien-1-yl trihydrogen diphosphate, (2Z)-2-fluoro-3,7-dimethylocta-2,6-dien-1-yl trihydrogen diphosphate, 2-ethyl-2-(hydroxymethyl)propane-1,3-diol, ... | | Authors: | Karuppiah, V, Leys, D, Scrutton, N.S. | | Deposit date: | 2017-05-09 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structural Basis of Catalysis in the Bacterial Monoterpene Synthases Linalool Synthase and 1,8-Cineole Synthase.
ACS Catal, 7, 2017
|
|
5NX5
 
 | | Crystal structure of Linalool/Nerolidol synthase from Streptomyces clavuligerus in complex with 2-fluorogeranyl diphosphate | | Descriptor: | (2Z)-2-fluoro-3,7-dimethylocta-2,6-dien-1-yl trihydrogen diphosphate, CHLORIDE ION, GLYCEROL, ... | | Authors: | Karuppiah, V, Leys, D, Scrutton, N.S. | | Deposit date: | 2017-05-09 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural Basis of Catalysis in the Bacterial Monoterpene Synthases Linalool Synthase and 1,8-Cineole Synthase.
ACS Catal, 7, 2017
|
|
5NX6
 
 | | Crystal structure of 1,8-cineole synthase from Streptomyces clavuligerus in complex with 2-fluoroneryl diphosphate | | Descriptor: | (2E)-2-fluoro-3,7-dimethylocta-2,6-dien-1-yl trihydrogen diphosphate, 2-ethyl-2-(hydroxymethyl)propane-1,3-diol, ETHYL DIMETHYL AMMONIO PROPANE SULFONATE, ... | | Authors: | Karuppiah, V, Leys, D, Scrutton, N.S. | | Deposit date: | 2017-05-09 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural Basis of Catalysis in the Bacterial Monoterpene Synthases Linalool Synthase and 1,8-Cineole Synthase.
ACS Catal, 7, 2017
|
|
2H3X
 
 | | Crystal Structure of an Electron Transfer Complex Between Aromatic Amine Dehydrogenase and Azurin from Alcaligenes Faecalis (Form 3) | | Descriptor: | Aromatic Amine Dehydrogenase, Azurin, COPPER (II) ION | | Authors: | Sukumar, N, Chen, Z, Leys, D, Scrutton, N.S, Ferrati, D, Merli, A, Rossi, G.L, Bellamy, H.D, Chistoserdov, A, Davidson, V.L, Mathews, F.S. | | Deposit date: | 2006-05-23 | | Release date: | 2006-11-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of an Electron Transfer Complex between Aromatic Amine Dehydrogenase and Azurin from Alcaligenes faecalis.
Biochemistry, 45, 2006
|
|
2IAA
 
 | | Crystal Structure of an Electron Transfer Complex Between Aromatic Amine Dephydrogenase and Azurin from Alcaligenes Faecalis (Form 2) | | Descriptor: | Aromatic Amine Dehydrogenase, Azurin, COPPER (II) ION | | Authors: | Sukumar, N, Chen, Z, Leys, D, Scrutton, N.S, Ferrati, D, Merli, A, Rossi, G.L, Bellamy, H.D, Chistoserdov, A, Davidson, V.L, Mathews, F.S. | | Deposit date: | 2006-09-07 | | Release date: | 2006-11-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of an Electron Transfer Complex between Aromatic Amine Dehydrogenase and Azurin from Alcaligenes faecalis.
Biochemistry, 45, 2006
|
|
2H47
 
 | | Crystal Structure of an Electron Transfer Complex Between Aromatic Amine Dephydrogenase and Azurin from Alcaligenes Faecalis (Form 1) | | Descriptor: | Aromatic Amine Dehydrogenase, Azurin, COPPER (II) ION | | Authors: | Sukumar, N, Chen, Z, Leys, D, Scrutton, N.S, Ferrati, D, Merli, A, Rossi, G.L, Bellamy, H.D, Chistoserdov, A, Davidson, V.L, Mathews, F.S. | | Deposit date: | 2006-05-23 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of an Electron Transfer Complex between Aromatic Amine Dehydrogenase and Azurin from Alcaligenes faecalis.
Biochemistry, 45, 2006
|
|
1GVQ
 
 | |
1VYR
 
 | | Structure of pentaerythritol tetranitrate reductase complexed with picric acid | | Descriptor: | FLAVIN MONONUCLEOTIDE, PENTAERYTHRITOL TETRANITRATE REDUCTASE, PICRIC ACID | | Authors: | Barna, T, Moody, P.C.E. | | Deposit date: | 2004-05-05 | | Release date: | 2004-06-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Atomic Resolution Structures and Solution Behavior of Enzyme-Substrate Complexes of Enterobacter Cloacae Pb2 Pentaerythritol Tetranitrate Reductase: Multiple Conformational States and Implications for the Mechanism of Nitroaromatic Explosive Degradation
J.Biol.Chem., 279, 2004
|
|
1VYP
 
 | | Structure of pentaerythritol tetranitrate reductase W102F mutant and complexed with picric acid | | Descriptor: | FLAVIN MONONUCLEOTIDE, PENTAERYTHRITOL TETRANITRATE REDUCTASE, PICRIC ACID | | Authors: | Barna, T, Moody, P.C.E. | | Deposit date: | 2004-05-04 | | Release date: | 2004-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Atomic Resolution Structures and Solution Behavior of Enzyme-Substrate Complexes of Enterobacter Cloacae Pb2 Pentaerythritol Tetranitrate Reductase: Multiple Conformational States and Implications for the Mechanism of Nitroaromatic Explosive Degradation
J.Biol.Chem., 279, 2004
|
|
1GES
 
 | |
