3EYD
 
 | | Structure of HCV NS3-4A Protease with an Inhibitor Derived from a Boronic Acid | | Descriptor: | HCV NS3, HCV NS4a peptide, ZINC ION, ... | | Authors: | Venkatraman, S, Wu, W, Prongay, A.J, Girijavallabhan, V, Njoroge, F.G. | | Deposit date: | 2008-10-20 | | Release date: | 2009-02-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Potent inhibitors of HCV-NS3 protease derived from boronic acids.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
5NL4
 
 | | Crystal structure of Zn1.3-E16V human ubiquitin (hUb) mutant adduct, from a solution 35 mM zinc acetate/1.3 mM E16V hUb | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Fermani, S, Falini, G. | | Deposit date: | 2017-04-04 | | Release date: | 2017-04-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Aggregation Pathways of Native-Like Ubiquitin Promoted by Single-Point Mutation, Metal Ion Concentration, and Dielectric Constant of the Medium.
Chemistry, 24, 2018
|
|
5NLI
 
 | | Crystal structure of Zn2-E16V human ubiquitin (hUb) mutant adduct, from a solution 35 mM zinc acetate/10% v/v TFE/1.3 mM E16V hUb | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Polyubiquitin-C, ... | | Authors: | Fermani, S, Falini, G. | | Deposit date: | 2017-04-04 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Aggregation Pathways of Native-Like Ubiquitin Promoted by Single-Point Mutation, Metal Ion Concentration, and Dielectric Constant of the Medium.
Chemistry, 24, 2018
|
|
7SWL
 
 | | CryoEM structure of the N-terminal-deleted Rix7 AAA-ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kocaman, S, Stanley, R.E, Lo, Y.H, Krahn, J, Dandey, V.P, Sobhany, M, Petrovich, M, Williams, J.G, Deterding, L.J, Borgnia, M.J, Etigunta, S. | | Deposit date: | 2021-11-20 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Communication network within the essential AAA-ATPase Rix7 drives ribosome assembly.
Pnas Nexus, 1, 2022
|
|
7T0V
 
 | | CryoEM structure of the crosslinked Rix7 AAA-ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kocaman, S, Stanley, R.E, Lo, Y.H, Krahn, J, Dandey, V.P, Sobhany, M, Petrovich, M, Williams, J.G, Deterding, L.J, Borgnia, M.J, Etigunta, S. | | Deposit date: | 2021-11-30 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Communication network within the essential AAA-ATPase Rix7 drives ribosome assembly.
Pnas Nexus, 1, 2022
|
|
5NMC
 
 | |
5NL5
 
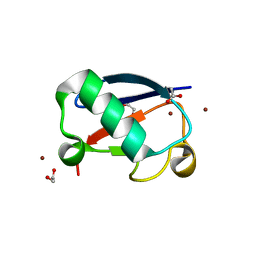 | | Crystal structure of Zn1.7-E16V human ubiquitin (hUb) mutant adduct, from a solution 70 mM zinc acetate/1.3 mM E16V hUb | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Polyubiquitin-B, ... | | Authors: | Fermani, S, Falini, G. | | Deposit date: | 2017-04-04 | | Release date: | 2017-04-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Aggregation Pathways of Native-Like Ubiquitin Promoted by Single-Point Mutation, Metal Ion Concentration, and Dielectric Constant of the Medium.
Chemistry, 24, 2018
|
|
5NLF
 
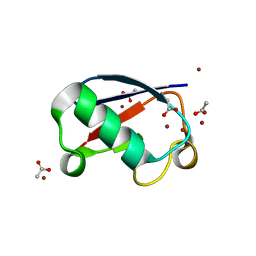 | | Crystal structure of Zn2.7-E16V human ubiquitin (hUb) mutant adduct, from a solution 100 mM zinc acetate/1.3 mM E16V hUb | | Descriptor: | ACETATE ION, Polyubiquitin-C, ZINC ION | | Authors: | Fermani, S, Falini, G. | | Deposit date: | 2017-04-04 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Aggregation Pathways of Native-Like Ubiquitin Promoted by Single-Point Mutation, Metal Ion Concentration, and Dielectric Constant of the Medium.
Chemistry, 24, 2018
|
|
5NLJ
 
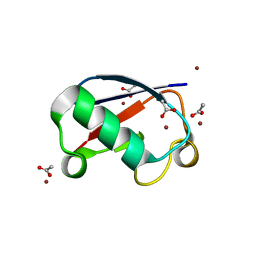 | | Crystal structure of Zn3-E16V human ubiquitin (hUb) mutant adduct, from a solution 70 mM zinc acetate/20% v/v TFE/1.3 mM E16V hUb | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Fermani, S, Falini, G. | | Deposit date: | 2017-04-04 | | Release date: | 2017-04-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Aggregation Pathways of Native-Like Ubiquitin Promoted by Single-Point Mutation, Metal Ion Concentration, and Dielectric Constant of the Medium.
Chemistry, 24, 2018
|
|
6AIE
 
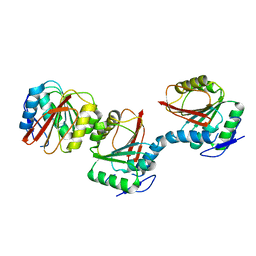 | | Crystal structure of a new form of RsmD-like RNA methyl transferase from Mycobacterium tuberculosis determined at 1.74 A resolution | | Descriptor: | Putative methyltransferase | | Authors: | Venkataraman, S, Dhankar, A, Sinha, K.M, Manivasakan, P, Iqbal, N, Singh, T.P, Prasad, B.V.L.S. | | Deposit date: | 2018-08-22 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of a new form of RsmD-like RNA methyl transferase from Mycobacterium tuberculosis determined at 1.74 A resolution
To Be Published
|
|
7UW7
 
 | | Crystal structure of the Human TRIP12 WWE domain (isoform 2) in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Isoform 2 of E3 ubiquitin-protein ligase TRIP12, UNKNOWN ATOM OR ION | | Authors: | Kimani, S, Dong, A, Li, Y, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-05-02 | | Release date: | 2022-06-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Crystal structure of the Human TRIP12 WWE domain (isoform 2) in complex with ADP
To Be Published
|
|
5MDU
 
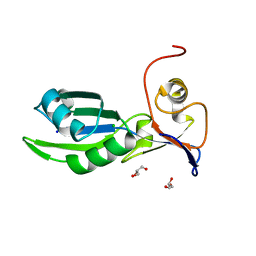 | | Structure of the RNA recognition motif (RRM) of Seb1 from S. pombe. | | Descriptor: | CHLORIDE ION, GLYCEROL, Rpb7-binding protein seb1, ... | | Authors: | Wittmann, S, Renner, M, El Omari, K, Adams, O, Vasiljeva, L, Grimes, J. | | Deposit date: | 2016-11-13 | | Release date: | 2017-04-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | The conserved protein Seb1 drives transcription termination by binding RNA polymerase II and nascent RNA.
Nat Commun, 8, 2017
|
|
7T3I
 
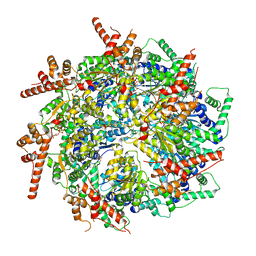 | | CryoEM structure of the Rix7 D2 Walker B mutant | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, PHOSPHATE ION, Rix7, ... | | Authors: | Kocaman, S, Stanley, R.E. | | Deposit date: | 2021-12-08 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Communication network within the essential AAA-ATPase Rix7 drives ribosome assembly.
Pnas Nexus, 1, 2022
|
|
5MDT
 
 | |
6WJL
 
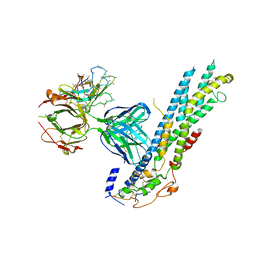 | | Crystal structure of Glypican-2 core protein in complex with D3 Fab | | Descriptor: | D3 Fab Heavy chain, D3 Fab Light Chain, Glypican-2, ... | | Authors: | Raman, S, Maris, J.M, Bosse, K.R, Julien, J.P. | | Deposit date: | 2020-04-14 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A GPC2 antibody-drug conjugate is efficacious against neuroblastoma and small-cell lung cancer via binding a conformational epitope.
Cell Rep Med, 2, 2021
|
|
4BPF
 
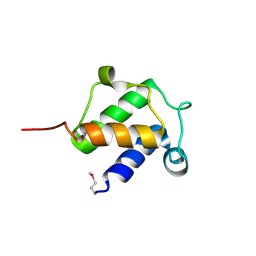 | |
4BPH
 
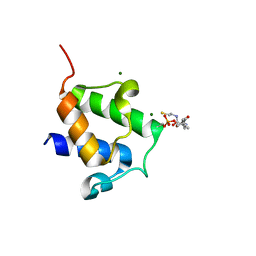 | | High resolution crystal structure of Bacillus subtilis DltC | | Descriptor: | 4'-PHOSPHOPANTETHEINE, D-ALANINE--POLY(PHOSPHORIBITOL) LIGASE SUBUNIT 2, MAGNESIUM ION | | Authors: | Zimmermann, S, Neumann, P, Stubbs, M.T. | | Deposit date: | 2013-05-26 | | Release date: | 2014-06-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-Resolution Structures of the D-Alanyl Carrier Protein (Dcp) Dltc from Bacillus Subtilis Reveal Equivalent Conformations of Apo- and Holo-Forms
FEBS Lett., 589, 2015
|
|
8T5I
 
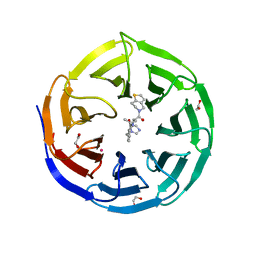 | | Crystal structure of human WDR5 in complex with MR4397 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, N-[(2S)-1-(6,7-dihydrothieno[3,2-c]pyridin-5(4H)-yl)-1-oxopentan-2-yl]-3-[(1H-imidazol-1-yl)methyl]benzamide, ... | | Authors: | Kimani, S, Dong, A, Li, F, Loppnau, P, Ackloo, S, Vedadi, M, Brown, P.J, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-06-13 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of human WDR5 in complex with MR4397
To be published
|
|
8F8E
 
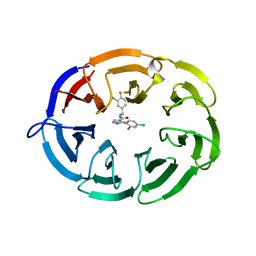 | | Crystal structure of the WDR domain of human DCAF1 in complex with OICR-8268 compound | | Descriptor: | (3P)-N-[(1S)-3-amino-1-(3-chloro-4-fluorophenyl)-3-oxopropyl]-3-(4-chloro-2-fluorophenyl)-1H-pyrazole-4-carboxamide, CITRIC ACID, DDB1- and CUL4-associated factor 1 | | Authors: | Kimani, S, Li, A, Dong, A, Li, Y, Hutchinson, A, Seitova, A, Wilson, B, Al-Awar, R, Vedadi, M, Brown, P, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-11-21 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of the WDR domain of human DCAF1 in complex with OICR-8268 compound
To be published
|
|
8ELD
 
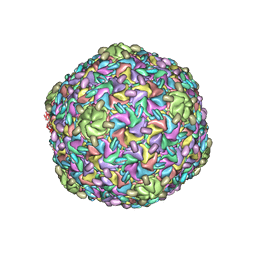 | |
8EM6
 
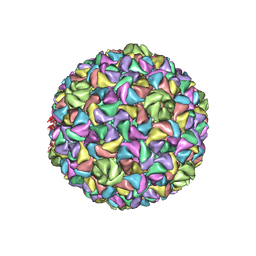 | |
8ES4
 
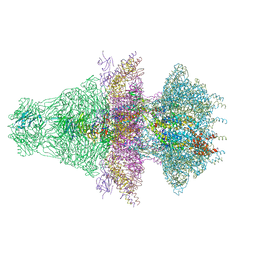 | |
6FHS
 
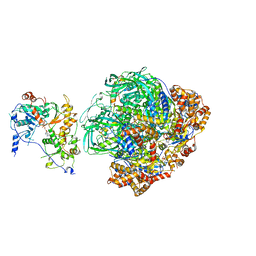 | | CryoEM Structure of INO80core | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Arp5, ... | | Authors: | Eustermann, S, Schall, K, Kostrewa, D, Strauss, M, Hopfner, K. | | Deposit date: | 2018-01-15 | | Release date: | 2018-04-25 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.754 Å) | | Cite: | Structural basis for ATP-dependent chromatin remodelling by the INO80 complex.
Nature, 556, 2018
|
|
5NUI
 
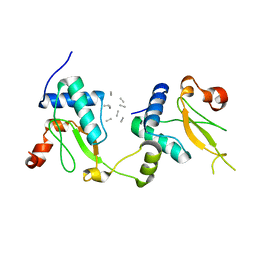 | | Crystal structure of SIVmac239 Nef in an ExxxLM endocytic sorting motif bound state | | Descriptor: | Protein Nef, SER-GLN-ILE-LYS-ARG-LEU-LEU-SER | | Authors: | Manrique, S, Horenkamp, F.A, Anand, K, Geyer, M. | | Deposit date: | 2017-04-30 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Endocytic sorting motif interactions involved in Nef-mediated downmodulation of CD4 and CD3.
Nat Commun, 8, 2017
|
|
6FML
 
 | | CryoEM Structure INO80core Nucleosome complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin related protein 5, ... | | Authors: | Eustermann, S, Schall, K, Kostrewa, D, Strauss, M, Hopfner, K. | | Deposit date: | 2018-01-31 | | Release date: | 2018-04-25 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.34 Å) | | Cite: | Structural basis for ATP-dependent chromatin remodelling by the INO80 complex.
Nature, 556, 2018
|
|
