2N75
 
 | | Solution NMR Structure of De novo designed protein, Rossmann2x2 Fold, Northeast Structural Genomics Consortium (NESG) Target OR446 | | Descriptor: | De novo designed protein | | Authors: | Liu, G, Lin, Y, Koga, N, Koga, R, Xiao, R, Janjua, H, Pederson, K, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-09-03 | | Release date: | 2016-01-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of De novo designed protein, Rossmann2x2 Fold, Northeast Structural Genomics Consortium (NESG) Target OR446
To be Published
|
|
3EAO
 
 | | Crystal structure of recombinant rat selenoprotein thioredoxin reductase 1 with oxidized C-terminal tail | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Thioredoxin reductase 1, ... | | Authors: | Sandalova, T, Cheng, Q, Lindqvist, Y, Arner, E. | | Deposit date: | 2008-08-26 | | Release date: | 2008-12-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure and catalysis of the selenoprotein thioredoxin reductase 1.
J.Biol.Chem., 284, 2009
|
|
3EAN
 
 | | Crystal structure of recombinant rat selenoprotein thioredoxin reductase 1 with reduced C-terminal tail | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Thioredoxin reductase 1 | | Authors: | Sandalova, T, Cheng, Q, Lindqvist, Y, Arner, E. | | Deposit date: | 2008-08-26 | | Release date: | 2008-12-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure and catalysis of the selenoprotein thioredoxin reductase 1.
J.Biol.Chem., 284, 2009
|
|
7PP7
 
 | | Thunberia alata 16:0-ACP desaturase | | Descriptor: | Acyl-[acyl-carrier-protein] 6-desaturase, FE (III) ION | | Authors: | Guy, J.E, Whittle, E, Cai, Y, Chai, J, Lindqvist, Y, Shanklin, J. | | Deposit date: | 2021-09-13 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Regioselectivity mechanism of the Thunbergia alata Delta 6-16:0-acyl carrier protein desaturase.
Plant Physiol., 188, 2022
|
|
1AL8
 
 | |
1AL7
 
 | |
1BS1
 
 | | DETHIOBIOTIN SYNTHETASE COMPLEXED WITH DETHIOBIOTIN, ADP , INORGANIC PHOSPHATE AND MAGNESIUM | | Descriptor: | 8-AMINO-7-CARBOXYAMINO-NONANOIC ACID WITH ALUMINUM FLUORIDE, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kaeck, H, Sandmark, J, Gibson, K.J, Schneider, G, Lindqvist, Y. | | Deposit date: | 1998-08-31 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of two quaternary complexes of dethiobiotin synthetase, enzyme-MgADP-AlF3-diaminopelargonic acid and enzyme-MgADP-dethiobiotin-phosphate; implications for catalysis.
Protein Sci., 7, 1998
|
|
3KG1
 
 | | Crystal structure of SnoaB, a cofactor-independent oxygenase from Streptomyces nogalater, mutant N63A | | Descriptor: | CHLORIDE ION, SnoaB | | Authors: | Koskiniemi, H, Grocholski, T, Lindqvist, Y, Mantsala, P, Niemi, J, Schneider, G. | | Deposit date: | 2009-10-28 | | Release date: | 2010-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the cofactor-independent monooxygenase SnoaB from Streptomyces nogalater: implications for the reaction mechanism
Biochemistry, 49, 2010
|
|
3KG0
 
 | | Crystal structure of SnoaB, a cofactor-independent oxygenase from Streptomyces nogalater, determined to 1.7 resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SnoaB | | Authors: | Koskiniemi, H, Grocholski, T, Lindqvist, Y, Mantsala, P, Niemi, J, Schneider, G. | | Deposit date: | 2009-10-28 | | Release date: | 2010-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the cofactor-independent monooxygenase SnoaB from Streptomyces nogalater: implications for the reaction mechanism
Biochemistry, 49, 2010
|
|
3KNG
 
 | | Crystal structure of SnoaB, a cofactor-independent oxygenase from Streptomyces nogalater, determined to 1.9 resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SULFATE ION, ... | | Authors: | Koskiniemi, H, Grocholski, T, Lindqvist, Y, Mantsala, P, Niemi, J, Schneider, G. | | Deposit date: | 2009-11-12 | | Release date: | 2010-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the cofactor-independent monooxygenase SnoaB from Streptomyces nogalater: implications for the reaction mechanism
Biochemistry, 49, 2010
|
|
3LCP
 
 | | Crystal structure of the carbohydrate recognition domain of LMAN1 in complex with MCFD2 | | Descriptor: | CALCIUM ION, Multiple coagulation factor deficiency protein 2, Protein ERGIC-53 | | Authors: | Wigren, E, Bourhis, J.M, Kursula, I, Guy, J.E, Lindqvist, Y. | | Deposit date: | 2010-01-11 | | Release date: | 2010-01-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of the LMAN1-CRD/MCFD2 transport receptor complex provides insight into combined deficiency of factor V and factor VIII.
Febs Lett., 584, 2010
|
|
1BII
 
 | | THE CRYSTAL STRUCTURE OF H-2DD MHC CLASS I IN COMPLEX WITH THE HIV-1 DERIVED PEPTIDE P18-110 | | Descriptor: | BETA-2 MICROGLOBULIN, DECAMERIC PEPTIDE, MHC CLASS I H-2DD | | Authors: | Achour, A, Persson, K, Harris, R.A, Sundback, J, Sentman, C.L, Lindqvist, Y, Schneider, G, Karre, K. | | Deposit date: | 1998-06-11 | | Release date: | 1998-10-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of H-2Dd MHC class I complexed with the HIV-1-derived peptide P18-I10 at 2.4 A resolution: implications for T cell and NK cell recognition.
Immunity, 9, 1998
|
|
6STD
 
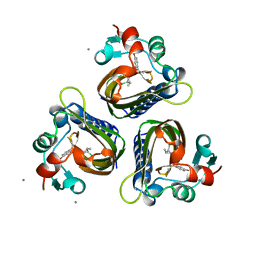 | | SCYTALONE DEHYDRATASE PLUS INHIBITOR 3 | | Descriptor: | 2,2-DICHLORO-1-METHANESULFINYL-3-METHYL-CYCLOPROPANECARBOXYLIC ACID [1-(4-BROMO-PHENYL)-ETHYL]-AMIDE, CALCIUM ION, Scytalone dehydratase | | Authors: | Wawrzak, Z, Sandalova, T, Steffens, J.J, Basarab, G.S, Lundqvist, T, Lindqvist, Y, Jordan, D.B. | | Deposit date: | 1999-02-11 | | Release date: | 1999-12-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution structures of scytalone dehydratase-inhibitor complexes crystallized at physiological pH.
Proteins, 35, 1999
|
|
5EPA
 
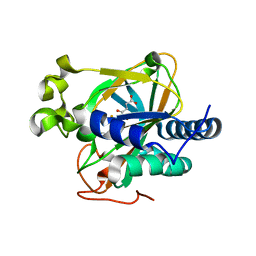 | | Crystal structure of non-heme alpha ketoglutarate dependent carbocyclase SnoK from nogalamycin biosynthesis | | Descriptor: | 2-OXOGLUTARIC ACID, FE (III) ION, MAGNESIUM ION, ... | | Authors: | Selvaraj, B, Lindqvist, Y, Siitonen, V, Niiranen, L, Metsa-Ketela, M, Schneider, G. | | Deposit date: | 2015-11-11 | | Release date: | 2016-05-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Divergent non-heme iron enzymes in the nogalamycin biosynthetic pathway.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1NGS
 
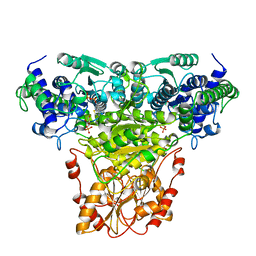 | | COMPLEX OF TRANSKETOLASE WITH THIAMIN DIPHOSPHATE, CA2+ AND ACCEPTOR SUBSTRATE ERYTHROSE-4-PHOSPHATE | | Descriptor: | CALCIUM ION, ERYTHOSE-4-PHOSPHATE, THIAMINE DIPHOSPHATE, ... | | Authors: | Nilsson, U, Lindqvist, Y, Schneider, G. | | Deposit date: | 1996-09-25 | | Release date: | 1997-02-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Examination of substrate binding in thiamin diphosphate-dependent transketolase by protein crystallography and site-directed mutagenesis.
J.Biol.Chem., 272, 1997
|
|
5ERL
 
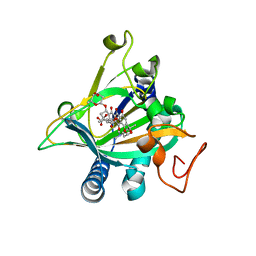 | | Crystal structure of the epimerase SnoN in complex with Ni2+, succinate and nogalamycin RO | | Descriptor: | NICKEL (II) ION, Nogalamycin RO, SUCCINIC ACID, ... | | Authors: | Selvaraj, B, Lindqvist, Y, Siitonen, V, Metsa-Ketela, M, Schneider, G. | | Deposit date: | 2015-11-14 | | Release date: | 2016-05-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Divergent non-heme iron enzymes in the nogalamycin biosynthetic pathway.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5EP9
 
 | | Crystal structure of the non-heme alpha ketoglutarate dependent epimerase SnoN from nogalamycin biosynthesis | | Descriptor: | 2-OXOGLUTARIC ACID, ACETATE ION, FE (III) ION, ... | | Authors: | Selvaraj, B, Lindqvist, Y, Niiranen, L, Siitonen, V, Metsa-Ketela, M, Schneider, G. | | Deposit date: | 2015-11-11 | | Release date: | 2016-05-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Divergent non-heme iron enzymes in the nogalamycin biosynthetic pathway.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5EZ7
 
 | | Crystal structure of the FAD dependent oxidoreductase PA4991 from Pseudomonas aeruginosa | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MERCURY (II) ION, flavoenzyme PA4991 | | Authors: | Jacewicz, A, Schnell, R, Lindqvist, Y, Schneider, G. | | Deposit date: | 2015-11-26 | | Release date: | 2016-02-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the flavoenzyme PA4991 from Pseudomonas aeruginosa.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5EQU
 
 | | Crystal structure of the epimerase SnoN in complex with Fe3+, alpha ketoglutarate and nogalamycin RO | | Descriptor: | 2-OXOGLUTARIC ACID, FE (III) ION, Nogalamycin RO, ... | | Authors: | Selvaraj, B, Lindqvist, Y, Siitonen, V, Metsa-Ketela, M, Schneider, G. | | Deposit date: | 2015-11-13 | | Release date: | 2016-05-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Divergent non-heme iron enzymes in the nogalamycin biosynthetic pathway.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1AHJ
 
 | | NITRILE HYDRATASE | | Descriptor: | FE (III) ION, NITRILE HYDRATASE (SUBUNIT ALPHA), NITRILE HYDRATASE (SUBUNIT BETA) | | Authors: | Huang, W, Schneider, G, Lindqvist, Y. | | Deposit date: | 1997-04-05 | | Release date: | 1998-04-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of nitrile hydratase reveals a novel iron centre in a novel fold.
Structure, 5, 1997
|
|
1A82
 
 | | DETHIOBIOTIN SYNTHETASE FROM ESCHERICHIA COLI, COMPLEX WITH SUBSTRATES ATP AND DIAMINOPELARGONIC ACID | | Descriptor: | 7,8-DIAMINO-NONANOIC ACID, ADENOSINE-5'-TRIPHOSPHATE, DETHIOBIOTIN SYNTHETASE, ... | | Authors: | Kaeck, H, Gibson, K.J, Lindqvist, Y, Schneider, G. | | Deposit date: | 1998-03-31 | | Release date: | 1999-05-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Snapshot of a phosphorylated substrate intermediate by kinetic crystallography.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1VGR
 
 | | Formyl-CoA transferase mutant Asp169 to Glu | | Descriptor: | COENZYME A, Formyl-coenzyme A transferase | | Authors: | Ricagno, S, Jonsson, S, Richards, N.G, Lindqvist, Y. | | Deposit date: | 2004-04-28 | | Release date: | 2004-08-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Kinetic and mechanistic characterization of the formyl-CoA transferase from Oxalobacter formigenes
J.Biol.Chem., 279, 2004
|
|
1ONR
 
 | | STRUCTURE OF TRANSALDOLASE B | | Descriptor: | TRANSALDOLASE B | | Authors: | Jia, J, Huang, W, Lindqvist, Y, Schneider, G. | | Deposit date: | 1996-08-13 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structure of transaldolase B from Escherichia coli suggests a circular permutation of the alpha/beta barrel within the class I aldolase family.
Structure, 4, 1996
|
|
1B3N
 
 | | BETA-KETOACYL CARRIER PROTEIN SYNTHASE AS A DRUG TARGET, IMPLICATIONS FROM THE CRYSTAL STRUCTURE OF A COMPLEX WITH THE INHIBITOR CERULENIN. | | Descriptor: | (2S, 3R)-3-HYDROXY-4-OXO-7,10-TRANS,TRANS-DODECADIENAMIDE, PROTEIN (KETOACYL ACYL CARRIER PROTEIN SYNTHASE 2) | | Authors: | Moche, M, Schneider, G, Edwards, P, Dehesh, K, Lindqvist, Y. | | Deposit date: | 1998-12-14 | | Release date: | 1999-04-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of the complex between the antibiotic cerulenin and its target, beta-ketoacyl-acyl carrier protein synthase.
J.Biol.Chem., 274, 1999
|
|
1VGQ
 
 | | Formyl-CoA transferase mutant Asp169 to Ala | | Descriptor: | Formyl-coenzyme A transferase, OXIDIZED COENZYME A | | Authors: | Ricagno, S, Jonsson, S, Richards, N.G, Lindqvist, Y. | | Deposit date: | 2004-04-28 | | Release date: | 2004-08-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Kinetic and mechanistic characterization of the formyl-CoA transferase from Oxalobacter formigenes
J.Biol.Chem., 279, 2004
|
|
