2GKO
 
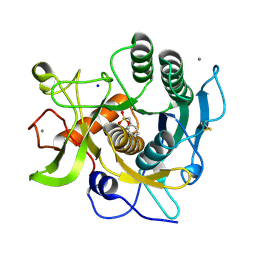 | | S41 Psychrophilic Protease | | Descriptor: | CALCIUM ION, SODIUM ION, microbial serine proteinases; subtilisin, ... | | Authors: | Walter, R.L, Mekel, M.J, Grayling, R.A, Arnold, F.H, Wintrode, P.L, Almog, O. | | Deposit date: | 2006-04-03 | | Release date: | 2007-05-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The crystal structures of the psychrophilic subtilisin S41 and the mesophilic subtilisin Sph reveal the same calcium-loaded state.
Proteins, 74, 2009
|
|
3D43
 
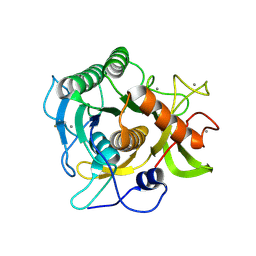 | | The crystal structure of Sph at 0.8A | | Descriptor: | CALCIUM ION, Sphericase | | Authors: | Almog, O. | | Deposit date: | 2008-05-13 | | Release date: | 2009-04-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | The crystal structures of the psychrophilic subtilisin S41 and the mesophilic subtilisin Sph reveal the same calcium-loaded state.
Proteins, 74, 2009
|
|
7MW6
 
 | |
7MW4
 
 | |
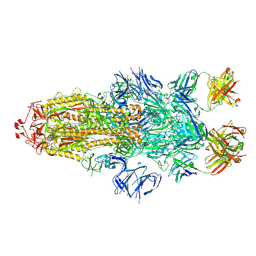
7MW2
 
 | |
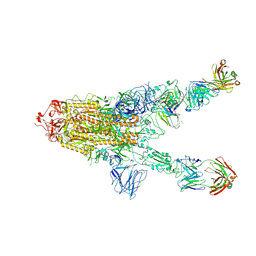
7MW5
 
 | |
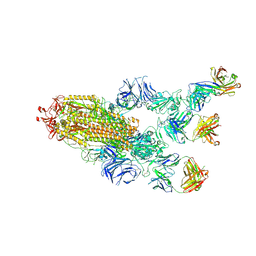
7MW3
 
 | |
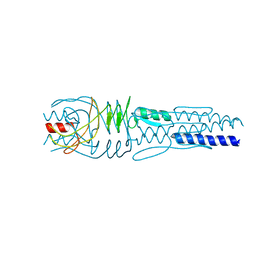
3LAA
 
 | |
3LA9
 
 | |
1A6T
 
 | |
1XZ9
 
 | | Structure of AF-6 PDZ domain | | Descriptor: | Afadin | | Authors: | Joshi, M, Boisguerin, P, Leitner, D, Volkmer-Engert, R, Moelling, K, Schade, M, Schmieder, P, Krause, G, Oschkinat, H. | | Deposit date: | 2004-11-12 | | Release date: | 2005-11-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Discovery of low-molecular-weight ligands for the AF6 PDZ domain.
Angew.Chem.Int.Ed.Engl., 45, 2006
|
|
5DCL
 
 | | Structure of a lantibiotic response regulator: N terminal domain of the nisin resistance regulator NsrR | | Descriptor: | 1,2-ETHANEDIOL, PhoB family transcriptional regulator | | Authors: | Khosa, S, Kleinschrodt, D, Hoeppner, A, Smits, S.H. | | Deposit date: | 2015-08-24 | | Release date: | 2016-03-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structure of the Response Regulator NsrR from Streptococcus agalactiae, Which Is Involved in Lantibiotic Resistance.
Plos One, 11, 2016
|
|
5DCM
 
 | | Structure of a lantibiotic response regulator: C-terminal domain of the nisin resistance regulator NsrR | | Descriptor: | PhoB family transcriptional regulator | | Authors: | Khosa, S, Kleinschrodt, D, Hoeppner, A, Smits, S.H.J. | | Deposit date: | 2015-08-24 | | Release date: | 2016-07-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the Response Regulator NsrR from Streptococcus agalactiae, Which Is Involved in Lantibiotic Resistance.
Plos One, 11, 2016
|
|
1SHX
 
 | | Ephrin A5 ligand structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ephrin-A5 | | Authors: | Himanen, J.P, Barton, W.A, Nikolov, D.B, Jeffrey, P.D. | | Deposit date: | 2004-02-26 | | Release date: | 2005-04-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Three distinct molecular surfaces in ephrin-A5 are essential for a functional interaction with EphA3.
J.Biol.Chem., 280, 2005
|
|
7ZIH
 
 | | Crystal structure of human tryptophan hydroxylase 1 in complex with inhibitor AG-01-128 | | Descriptor: | 8-(1~{H}-benzimidazol-2-ylmethyl)-3-ethyl-7-(phenylmethyl)purine-2,6-dione, DI(HYDROXYETHYL)ETHER, FE (III) ION, ... | | Authors: | Schuetz, A, Gogolin, A, Pfeifer, J, Mallow, K, Nazare, M, Specker, E, Heinemann, U. | | Deposit date: | 2022-04-08 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.46890831 Å) | | Cite: | Structure-Based Design of Xanthine-Benzimidazole Derivatives as Novel and Potent Tryptophan Hydroxylase Inhibitors.
J.Med.Chem., 65, 2022
|
|
7ZII
 
 | | Crystal structure of human tryptophan hydroxylase 1 in complex with inhibitor KM-05-193 | | Descriptor: | 8-(5~{H}-[1,3]dioxolo[4,5-f]benzimidazol-6-ylmethyl)-7-(phenylmethyl)-3-propyl-purine-2,6-dione, FE (III) ION, GLYCEROL, ... | | Authors: | Schuetz, A, Mallow, K, Nazare, M, Specker, E, Heinemann, U. | | Deposit date: | 2022-04-08 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6280005 Å) | | Cite: | Structure-Based Design of Xanthine-Benzimidazole Derivatives as Novel and Potent Tryptophan Hydroxylase Inhibitors.
J.Med.Chem., 65, 2022
|
|
7ZIK
 
 | | Crystal structure of human tryptophan hydroxylase 1 in complex with inhibitor LP533401 | | Descriptor: | (2~{R})-2-azanyl-3-[4-[2-azanyl-6-[(1~{R})-1-[4-chloranyl-2-(3-methylpyrazol-1-yl)phenyl]-2,2,2-tris(fluoranyl)ethoxy]pyrimidin-4-yl]phenyl]propanoic acid, FE (III) ION, Tryptophan 5-hydroxylase 1 | | Authors: | Schuetz, A, Heinemann, U. | | Deposit date: | 2022-04-08 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.58925915 Å) | | Cite: | Structure-Based Design of Xanthine-Benzimidazole Derivatives as Novel and Potent Tryptophan Hydroxylase Inhibitors.
J.Med.Chem., 65, 2022
|
|
7ZIJ
 
 | | Crystal structure of human tryptophan hydroxylase 1 in complex with inhibitor KM-05-080 | | Descriptor: | 8-(1~{H}-benzimidazol-2-ylmethyl)-3-cyclopropyl-7-(phenylmethyl)purine-2,6-dione, FE (III) ION, Tryptophan 5-hydroxylase 1 | | Authors: | Schuetz, A, Mallow, K, Nazare, M, Specker, E, Heinemann, U. | | Deposit date: | 2022-04-08 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.94678366 Å) | | Cite: | Structure-Based Design of Xanthine-Benzimidazole Derivatives as Novel and Potent Tryptophan Hydroxylase Inhibitors.
J.Med.Chem., 65, 2022
|
|
7ZIG
 
 | | Crystal structure of human tryptophan hydroxylase 1 in complex with inhibitor KM-05-060 | | Descriptor: | (2~{R})-2-azanyl-5-[[2-[[3-methyl-2,6-bis(oxidanylidene)-7-(phenylmethyl)purin-8-yl]methyl]-1~{H}-benzimidazol-5-yl]amino]-5-oxidanylidene-pentanoic acid, FE (III) ION, Tryptophan 5-hydroxylase 1 | | Authors: | Schuetz, A, Mallow, K, Nazare, M, Specker, E, Heinemann, U. | | Deposit date: | 2022-04-08 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.808885 Å) | | Cite: | Structure-Based Design of Xanthine-Benzimidazole Derivatives as Novel and Potent Tryptophan Hydroxylase Inhibitors.
J.Med.Chem., 65, 2022
|
|
7ZIF
 
 | | Crystal structure of human tryptophan hydroxylase 1 in complex with inhibitor KM-480 | | Descriptor: | (2R)-2-azanyl-5-[[2-[3-methyl-2,6-bis(oxidanylidene)-7-(phenylmethyl)purin-8-yl]sulfanyl-3H-benzimidazol-5-yl]amino]-5-oxidanylidene-pentanoic acid, FE (III) ION, GLYCEROL, ... | | Authors: | Schuetz, A, Ziebart, N, Weise, M, Mallow, K, Pfeifer, J, Nazare, M, Specker, E, Heinemann, U. | | Deposit date: | 2022-04-08 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.86859715 Å) | | Cite: | Structure-Based Design of Xanthine-Benzimidazole Derivatives as Novel and Potent Tryptophan Hydroxylase Inhibitors.
J.Med.Chem., 65, 2022
|
|
5NW5
 
 | | Crystal structure of the Rif1 N-terminal domain (RIF1-NTD) from Saccharomyces cerevisiae in complex with DNA | | Descriptor: | DNA (30-MER), DNA (60-MER), Telomere length regulator protein RIF1 | | Authors: | Bunker, R.D, Reinert, J.K, Shi, T, Thoma, N.H. | | Deposit date: | 2017-05-05 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (6.502 Å) | | Cite: | Rif1 maintains telomeres and mediates DNA repair by encasing DNA ends.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5NVR
 
 | |
3IFT
 
 | | Crystal structure of glycine cleavage system protein H from Mycobacterium tuberculosis, using X-rays from the Compact Light Source. | | Descriptor: | Glycine cleavage system H protein | | Authors: | Edwards, T.E, Abendroth, J, Staker, B, Mayer, C, Phan, I, Kelley, A, Analau, E, Leibly, D, Rifkin, J, Loewen, R, Ruth, R.D, Stewart, L.J, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D) | | Deposit date: | 2009-07-25 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure determination of the glycine cleavage system protein H of Mycobacterium tuberculosis using an inverse Compton synchrotron X-ray source.
J.Struct.Funct.Genom., 11, 2010
|
|
