5G0Q
 
 | | beta-glucuronidase with an activity-based probe (N-alkyl cyclophellitol aziridine) bound | | Descriptor: | (1R,2S,3R,4S,5S,6R)-7-(8-AZIDOOCTYL)-3,4,5-TRIHYDROXY-, BETA-GLUCURONIDASE, GLYCEROL | | Authors: | Jin, Y, Wu, L, Jiang, J.B, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2016-03-22 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Activity-based probes for functional interrogation of retaining beta-glucuronidases.
Nat. Chem. Biol., 13, 2017
|
|
5G0M
 
 | | beta-glucuronidase with an activity-based probe (N-acyl cyclophellitol aziridine) bound | | Descriptor: | (1S,2R,3R,4S,6S)-6-[(8-azidooctanoyl)amino]-2,3,4-trihydroxycyclohexane-1-carboxylate, BETA-GLUCURONIDASE, PHOSPHATE ION | | Authors: | Jin, Y, Wu, L, Jiang, J.B, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2016-03-21 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Structure of a Bacteria Beta-Glucuronidase in Gh79 Family
To be Published
|
|
5L77
 
 | | A glycoside hydrolase mutant with an unreacted activity based probe bound | | Descriptor: | (1~{R},2~{S},3~{R},4~{S},5~{S},6~{R})-7-[8-[(azanylidene-{4}-azanylidene)amino]octyl]-3,4,5-tris(oxidanyl)-7-azabicyclo[4.1.0]heptane-2-carboxylic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Jin, Y, Wu, L, Jiang, J.B, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2016-06-02 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Activity-based probes for functional interrogation of retaining beta-glucuronidases.
Nat. Chem. Biol., 13, 2017
|
|
3FCA
 
 | | Genetic Incorporation of a Metal-ion Chelating Amino Acid into proteins as biophysical probe | | Descriptor: | Cysteine synthase, ZINC ION | | Authors: | Wang, F, Lee, H, Spraggon, G, Schultz, P.G. | | Deposit date: | 2008-11-21 | | Release date: | 2009-02-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.149 Å) | | Cite: | Genetic incorporation of a metal-ion chelating amino acid into proteins as a biophysical probe.
J.Am.Chem.Soc., 131, 2009
|
|
1W01
 
 | |
1W02
 
 | |
8J9Q
 
 | | Crystal structure of UBR box of UBR4 apo | | Descriptor: | E3 ubiquitin-protein ligase UBR4, ZINC ION | | Authors: | Jeong, D.-E, KIm, S.-J, Shin, H.-C. | | Deposit date: | 2023-05-04 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Insights into the recognition mechanism in the UBR box of UBR4 for its specific substrates.
Commun Biol, 6, 2023
|
|
8J9R
 
 | |
8JOQ
 
 | |
8JOY
 
 | | Plk1 polo-box domain bound to HPV4 L2 residues 251-257 with pThr255 | | Descriptor: | Peptide from Minor capsid protein L2, Serine/threonine-protein kinase PLK1 | | Authors: | Ku, B, Jung, S. | | Deposit date: | 2023-06-09 | | Release date: | 2023-10-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal Structures of Plk1 Polo-Box Domain Bound to the Human Papillomavirus Minor Capsid Protein L2-Derived Peptide.
J.Microbiol, 61, 2023
|
|
8HNP
 
 | | Archaeal transcription factor Mutant | | Descriptor: | Archaeal transcription regulator | | Authors: | Bae, D.W, Cha, S.S. | | Deposit date: | 2022-12-08 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.39 Å) | | Cite: | An archaeal transcription factor EnfR with a novel 'eighth note' fold controls hydrogen production of a hyperthermophilic archaeon Thermococcus onnurineus NA1.
Nucleic Acids Res., 51, 2023
|
|
8HNO
 
 | | Archaeal transcription factor Wild type | | Descriptor: | Archaeal transcription regulator | | Authors: | Bae, D.W, Cha, S.S. | | Deposit date: | 2022-12-08 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | An archaeal transcription factor EnfR with a novel 'eighth note' fold controls hydrogen production of a hyperthermophilic archaeon Thermococcus onnurineus NA1.
Nucleic Acids Res., 51, 2023
|
|
3K1J
 
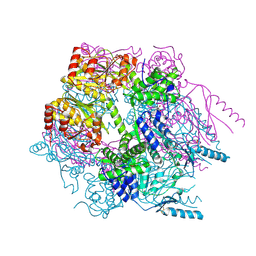 | | Crystal structure of Lon protease from Thermococcus onnurineus NA1 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Cha, S.S, An, Y.J. | | Deposit date: | 2009-09-28 | | Release date: | 2010-09-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Lon protease: molecular architecture of gated entry to a sequestered degradation chamber
Embo J., 29, 2010
|
|
1SMA
 
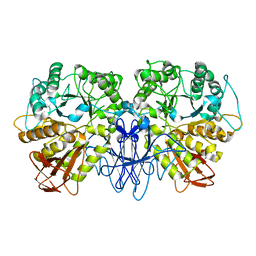 | | CRYSTAL STRUCTURE OF A MALTOGENIC AMYLASE | | Descriptor: | MALTOGENIC AMYLASE | | Authors: | Kim, J.S, Cha, S.S, Oh, B.H. | | Deposit date: | 1999-04-21 | | Release date: | 2000-04-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a maltogenic amylase provides insights into a catalytic versatility.
J.Biol.Chem., 274, 1999
|
|
1ZKJ
 
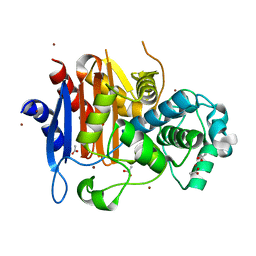 | | Structural Basis for the Extended Substrate Spectrum of CMY-10, a Plasmid-Encoded Class C beta-lactamase | | Descriptor: | ACETIC ACID, ZINC ION, extended-spectrum beta-lactamase | | Authors: | Cha, S.S, Jung, H.I, An, Y.J, Lee, S.H. | | Deposit date: | 2005-05-03 | | Release date: | 2006-04-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for the extended substrate spectrum of CMY-10, a plasmid-encoded class C beta-lactamase.
Mol.Microbiol., 60, 2006
|
|
5CBN
 
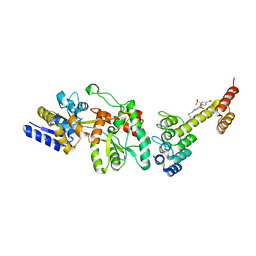 | | Fusion protein of mbp3-16 and B4 domain of protein A from staphylococcal aureus with chemical cross-linker EY-CBS | | Descriptor: | 2,2'-ethyne-1,2-diylbis{5-[(chloroacetyl)amino]benzenesulfonic acid}, Maltose-binding periplasmic protein, mbp3-16,Immunoglobulin G-binding protein A | | Authors: | Jeong, W.H, Lee, H, Song, D.H, Lee, J.O. | | Deposit date: | 2015-07-01 | | Release date: | 2016-03-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Connecting two proteins using a fusion alpha helix stabilized by a chemical cross linker.
Nat Commun, 7, 2016
|
|
5CBO
 
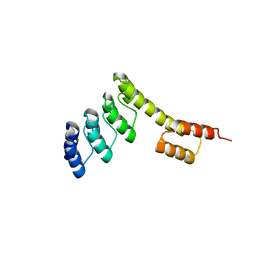 | | Fusion protein of mbp3-16 and B4 domain of protein A from staphylococcal aureus | | Descriptor: | mbp3-16,Immunoglobulin G-binding protein A | | Authors: | Jeong, W.H, Lee, H, Song, D.H, Lee, J.O. | | Deposit date: | 2015-07-01 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Connecting two proteins using a fusion alpha helix stabilized by a chemical cross linker.
Nat Commun, 7, 2016
|
|
5COC
 
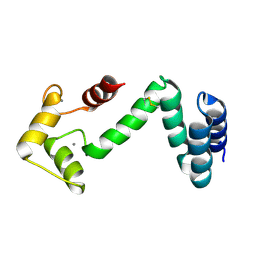 | | Fusion protein of human calmodulin and B4 domain of protein A from staphylococcal aureus | | Descriptor: | CALCIUM ION, Immunoglobulin G-binding protein A,Calmodulin | | Authors: | Jeong, W.H, Lee, H, Song, D.H, Lee, J.O. | | Deposit date: | 2015-07-20 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6691 Å) | | Cite: | Connecting two proteins using a fusion alpha helix stabilized by a chemical cross linker.
Nat Commun, 7, 2016
|
|
2MP8
 
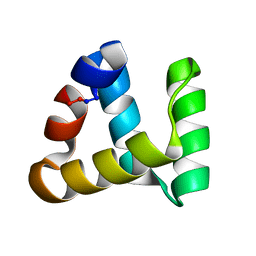 | | NMR structure of NKR-5-3B | | Descriptor: | NKR-5-3B | | Authors: | Rosengren, K.J, Craik, D.J. | | Deposit date: | 2014-05-13 | | Release date: | 2015-05-13 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Identification, Characterization, and Three-Dimensional Structure of the Novel Circular Bacteriocin, Enterocin NKR-5-3B, from Enterococcus faecium
Biochemistry, 54, 2015
|
|
5EWX
 
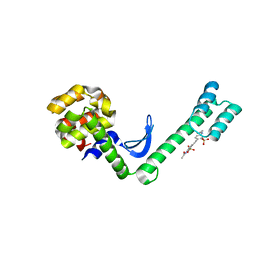 | | Fusion protein of T4 lysozyme and B4 domain of protein A from staphylococcal aureus with chemical cross-linker EY-CBS | | Descriptor: | 2,2'-ethyne-1,2-diylbis{5-[(chloroacetyl)amino]benzenesulfonic acid}, Endolysin,Immunoglobulin G-binding protein A,Endolysin | | Authors: | Jeong, W.H, Lee, H, Song, D.H, Lee, J.O. | | Deposit date: | 2015-11-22 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Connecting two proteins using a fusion alpha helix stabilized by a chemical cross linker.
Nat Commun, 7, 2016
|
|
6IWD
 
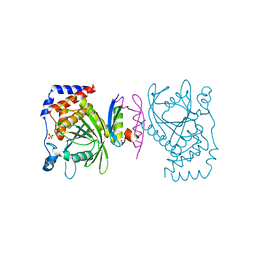 | | The PTP domain of human PTPN14 in a complex with the CR3 domain of HPV18 E7 | | Descriptor: | CHLORIDE ION, HPV18 E7, PHOSPHATE ION, ... | | Authors: | Yun, H.-Y, Kim, S.J, Ku, B. | | Deposit date: | 2018-12-05 | | Release date: | 2019-07-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for recognition of the tumor suppressor protein PTPN14 by the oncoprotein E7 of human papillomavirus.
Plos Biol., 17, 2019
|
|
1IZZ
 
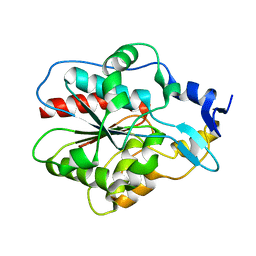 | | Crystal structure of Hsp31 | | Descriptor: | Hsp31 | | Authors: | Cha, S.S, Lee, S.J. | | Deposit date: | 2002-10-16 | | Release date: | 2003-10-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal structures of human DJ-1 and Escherichia coli Hsp31, which share an evolutionarily conserved domain
J.Biol.Chem., 278, 2003
|
|
1IZY
 
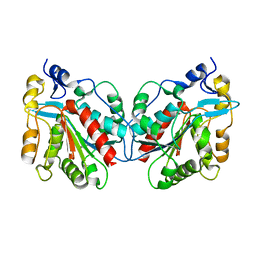 | | Crystal structure of Hsp31 | | Descriptor: | Hsp31 | | Authors: | Cha, S.S, Lee, S.J. | | Deposit date: | 2002-10-16 | | Release date: | 2003-10-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of human DJ-1 and Escherichia coli Hsp31, which share an evolutionarily conserved domain
J.Biol.Chem., 278, 2003
|
|
4IVK
 
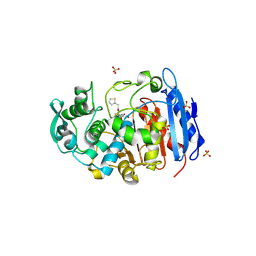 | | Crystal structure of a fammily VIII carboxylesterase in a complex with cephalothin. | | Descriptor: | CEPHALOTHIN GROUP, Carboxylesterases, SULFATE ION | | Authors: | An, Y.J, Kim, M.-K, Jeong, C.-S, Cha, S.-S. | | Deposit date: | 2013-01-23 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the beta-lactamase activity of EstU1, a family VIII carboxylesterase.
Proteins, 81, 2013
|
|
4IVI
 
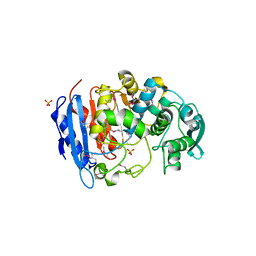 | | Crystal structure of a family VIII carboxylesterase. | | Descriptor: | Carboxylesterase, SULFATE ION | | Authors: | An, Y.J, Kim, M.-K, Jeong, C.-S, Cha, S.-S. | | Deposit date: | 2013-01-23 | | Release date: | 2013-06-19 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the beta-lactamase activity of EstU1, a family VIII carboxylesterase.
Proteins, 81, 2013
|
|
