3I7Z
 
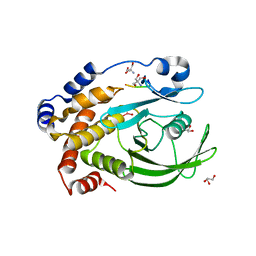 | | Protein Tyrosine Phosphatase 1B - Transition state analog for the first catalytic step | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, EGFR receptor fragment, GLYCEROL, ... | | Authors: | Brandao, T.A.S, Johnson, S.J, Hengge, A.C. | | Deposit date: | 2009-07-09 | | Release date: | 2010-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insights into the reaction of protein-tyrosine phosphatase 1B: crystal structures for transition state analogs of both catalytic steps.
J.Biol.Chem., 285, 2010
|
|
3I80
 
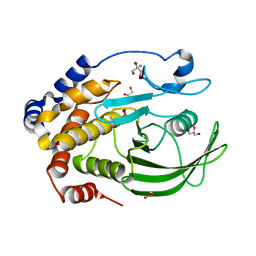 | | Protein Tyrosine Phosphatase 1B - Transition state analog for the second catalytic step | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Tyrosine-protein phosphatase non-receptor type 1, ... | | Authors: | Brandao, T.A.S, Johnson, S.J, Hengge, A.C. | | Deposit date: | 2009-07-09 | | Release date: | 2010-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Insights into the reaction of protein-tyrosine phosphatase 1B: crystal structures for transition state analogs of both catalytic steps.
J.Biol.Chem., 285, 2010
|
|
4ERC
 
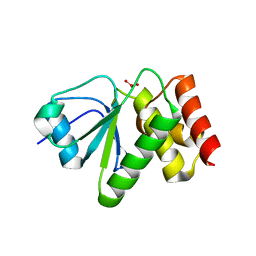 | | Structure of VHZ bound to metavanadate | | Descriptor: | Dual specificity protein phosphatase 23, oxido(dioxo)vanadium | | Authors: | Vyacheslav, K, Alvan, C.H, Sean, J.J. | | Deposit date: | 2012-04-19 | | Release date: | 2012-12-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | New Aspects of the Phosphatase VHZ Revealed by a High-Resolution Structure with Vanadate and Substrate Screening.
Biochemistry, 51, 2012
|
|
6YSF
 
 | |
6YSL
 
 | |
3PSJ
 
 | |
3PSK
 
 | |
3PSF
 
 | |
3TAP
 
 | |
3TAQ
 
 | |
3TAR
 
 | |
3TAN
 
 | |
3TI0
 
 | |
3THV
 
 | |
2YBY
 
 | | Structure of domains 6 and 7 of the mouse complement regulator Factor H | | Descriptor: | 1,2-ETHANEDIOL, COMPLEMENT FACTOR H | | Authors: | Everett, R.J, Caesar, J.J.E, Johnson, S.J, Tang, C.M, Lea, S.M. | | Deposit date: | 2011-03-30 | | Release date: | 2012-04-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Design and Evaluation of Meningococcal Vaccines Through Structure-Based Modification of Host and Pathogen Molecules.
Plos Pathog., 8, 2012
|
|
2YEQ
 
 | | Structure of PhoD | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ALKALINE PHOSPHATASE D, ... | | Authors: | Lillington, J.E.D, Rodriguez, F, Roversi, P, Johnson, S.J, Berks, B, Lea, S.M. | | Deposit date: | 2011-03-30 | | Release date: | 2012-04-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal Structure of the Bacillus Subtilis Phosphodiesterase Phod Reveals an Iron and Calcium-Containing Active Site.
J.Biol.Chem., 289, 2014
|
|
1U45
 
 | | 8oxoguanine at the pre-insertion site of the polymerase active site | | Descriptor: | DNA polymerase I, DNA primer strand, DNA template strand with 8-oxoguanine, ... | | Authors: | Hsu, G.W, Ober, M, Carell, T, Beese, L.S. | | Deposit date: | 2004-07-23 | | Release date: | 2004-09-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Error-prone replication of oxidatively damaged DNA by a high-fidelity DNA polymerase.
Nature, 431, 2004
|
|
1U4B
 
 | | Extension of an adenine-8oxoguanine mismatch | | Descriptor: | DNA polymerase I, DNA primer strand, DNA template strand with 8-oxoguanine, ... | | Authors: | Hsu, G.W, Ober, M, Carell, T, Beese, L.S. | | Deposit date: | 2004-07-23 | | Release date: | 2004-09-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Error-prone replication of oxidatively damaged DNA by a high-fidelity DNA polymerase.
Nature, 431, 2004
|
|
1UA0
 
 | | Aminofluorene DNA adduct at the pre-insertion site of a DNA polymerase | | Descriptor: | 2-AMINOFLUORENE, DNA polymerase I, DNA primer strand, ... | | Authors: | Hsu, G.W, Kiefer, J.R, Becherel, O.J, Fuchs, R.P.P, Beese, L.S. | | Deposit date: | 2004-08-11 | | Release date: | 2004-09-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Observing translesion synthesis of an aromatic amine DNA adduct by a high-fidelity DNA polymerase
J.Biol.Chem., 279, 2004
|
|
1UA1
 
 | | Structure of aminofluorene adduct paired opposite cytosine at the polymerase active site. | | Descriptor: | 2-AMINOFLUORENE, DNA polymerase I, DNA primer strand, ... | | Authors: | Hsu, G.W, Kiefer, J.R, Becherel, O.J, Fuchs, R.P.P, Beese, L.S. | | Deposit date: | 2004-08-11 | | Release date: | 2004-09-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Observing translesion synthesis of an aromatic amine DNA adduct by a high-fidelity DNA polymerase
J.Biol.Chem., 279, 2004
|
|
1U49
 
 | | Adenine-8oxoguanine mismatch at the polymerase active site | | Descriptor: | DNA polymerase I, DNA primer strand, DNA template strand with 8-oxoguanine, ... | | Authors: | Hsu, G.W, Ober, M, Carell, T, Beese, L.S. | | Deposit date: | 2004-07-23 | | Release date: | 2004-09-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Error-prone replication of oxidatively damaged DNA by a high-fidelity DNA polymerase.
Nature, 431, 2004
|
|
1U48
 
 | | Extension of a cytosine-8-oxoguanine base pair | | Descriptor: | DNA polymerase I, DNA primer strand, DNA template strand with 8-oxoguanine, ... | | Authors: | Hsu, G.W, Ober, M, Carell, T, Beese, L.S. | | Deposit date: | 2004-07-23 | | Release date: | 2004-09-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Error-prone replication of oxidatively damaged DNA by a high-fidelity DNA polymerase.
Nature, 431, 2004
|
|
1U47
 
 | | cytosine-8-Oxoguanine base pair at the polymerase active site | | Descriptor: | DNA polymerase I, DNA primer strand, DNA template strand with 8-oxoguanine, ... | | Authors: | Hsu, G.W, Ober, M, Carell, T, Beese, L.S. | | Deposit date: | 2004-07-23 | | Release date: | 2004-09-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Error-prone replication of oxidatively damaged DNA by a high-fidelity DNA polymerase.
Nature, 431, 2004
|
|
2HPH
 
 | |
1XC9
 
 | | Structure of a high-fidelity polymerase bound to a benzo[a]pyrene adduct that blocks replication | | Descriptor: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, DNA polymerase I, DNA primer strand, ... | | Authors: | Hsu, G.W, Huang, X, Luneva, N.P, Geacintov, N.E, Beese, L.S. | | Deposit date: | 2004-09-01 | | Release date: | 2004-12-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a High Fidelity DNA Polymerase Bound to a Benzo[a]pyrene Adduct That Blocks Replication
J.Biol.Chem., 280, 2005
|
|
