1EQM
 
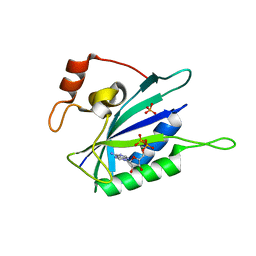 | | CRYSTAL STRUCTURE OF BINARY COMPLEX OF 6-HYDROXYMETHYL-7,8-DIHYDROPTERIN PYROPHOSPHOKINASE WITH ADENOSINE-5'-DIPHOSPHATE | | Descriptor: | 6-HYDROXYMETHYL-7,8-DIHYDROPTERIN PYROPHOSPHOKINASE, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Xiao, B, Blaszczyk, J, Ji, X. | | Deposit date: | 2000-04-05 | | Release date: | 2001-04-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Unusual conformational changes in 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase as revealed by X-ray crystallography and NMR.
J.Biol.Chem., 276, 2001
|
|
1EX8
 
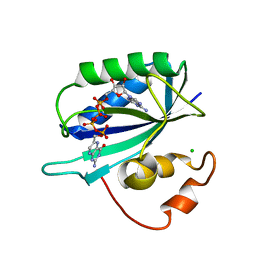 | | CRYSTAL STRUCTURE OF 6-HYDROXYMETHYL-7,8-DIHYDROPTERIN PYROPHOSPHOKINASE COMPLEXED WITH HP4A, THE TWO-SUBSTRATE-MIMICKING INHIBITOR | | Descriptor: | 6-(ADENOSINE TETRAPHOSPHATE-METHYL)-7,8-DIHYDROPTERIN, 6-HYDROXYMETHYL-7,8-DIHYDROPTERIN PYROPHOSPHOKINASE, CHLORIDE ION, ... | | Authors: | Blaszczyk, J, Ji, X. | | Deposit date: | 2000-05-01 | | Release date: | 2001-05-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Bisubstrate analogue inhibitors of 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase: synthesis and biochemical and crystallographic studies.
J.Med.Chem., 44, 2001
|
|
6AN6
 
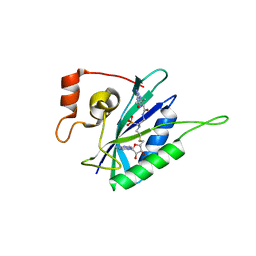 | | Crystal structure of Escherichia coli HPPK in complex with bisubstrate analogue inhibitor HP-72 | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, 5'-S-{2-[{2-[(2-amino-7,7-dimethyl-4-oxo-3,4,7,8-tetrahydropteridine-6-carbonyl)amino]ethyl}(phosphonomethyl)amino]ethyl}-5'-thioadenosine | | Authors: | Shaw, G.X, Shi, G, Ji, X. | | Deposit date: | 2017-08-12 | | Release date: | 2018-08-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bisubstrate analogue inhibitors of HPPK: Transition state mimetics
to be published
|
|
1F3B
 
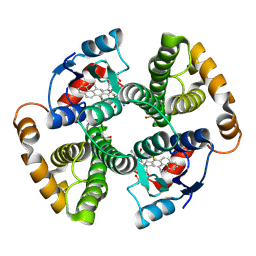 | | CRYSTAL STRUCTURE OF MGSTA1-1 IN COMPLEX WITH GLUTATHIONE CONJUGATE OF BENZO[A]PYRENE EPOXIDE | | Descriptor: | 2-AMINO-4-[1-(CARBOXYMETHYL-CARBAMOYL)-2-(9-HYDROXY-7,8-DIOXO-7,8,9,10-TETRAHYDRO-BENZO[DEF]CHRYSEN-10-YLSULFANYL)-ETHYLCARBAMOYL]-BUTYRIC ACID, GLUTATHIONE S-TRANSFERASE YA CHAIN | | Authors: | Gu, Y, Singh, S.V, Ji, X. | | Deposit date: | 2000-06-01 | | Release date: | 2000-10-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Residue R216 and catalytic efficiency of a murine class alpha glutathione S-transferase toward benzo[a]pyrene 7(R),8(S)-diol 9(S), 10(R)-epoxide.
Biochemistry, 39, 2000
|
|
1F3A
 
 | | CRYSTAL STRUCTURE OF MGSTA1-1 IN COMPLEX WITH GSH | | Descriptor: | GLUTATHIONE, GLUTATHIONE S-TRANSFERASE YA CHAIN | | Authors: | Gu, Y, Singh, S.V, Ji, X. | | Deposit date: | 2000-06-01 | | Release date: | 2000-10-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Residue R216 and catalytic efficiency of a murine class alpha glutathione S-transferase toward benzo[a]pyrene 7(R),8(S)-diol 9(S), 10(R)-epoxide.
Biochemistry, 39, 2000
|
|
1KU5
 
 | | Crystal Structure of recombinant histone HPhA from hyperthermophilic archaeon Pyrococcus horikoshii OT3 | | Descriptor: | ACETATE ION, HPhA, SULFATE ION | | Authors: | Li, T, Sun, F, Ji, X, Feng, Y, Rao, Z. | | Deposit date: | 2002-01-21 | | Release date: | 2003-08-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure based hyperthermostability of archaeal histone HPhA from Pyrococcus horikoshii
J.MOL.BIOL., 325, 2003
|
|
5JCU
 
 | |
3KUG
 
 | |
3KUE
 
 | | Crystal structure of E. coli HPPK(E77A) | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, ACETATE ION, CHLORIDE ION | | Authors: | Blaszczyk, J, Li, Y, Yan, H, Ji, X. | | Deposit date: | 2009-11-27 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.538 Å) | | Cite: | Roles of residues E77 and H115 in E. coli HPPK
To be Published
|
|
1RC5
 
 | |
5JMT
 
 | | Crystal structure of Zika virus NS3 helicase | | Descriptor: | NS3 helicase | | Authors: | Tian, H, Ji, X, Yang, X, Xie, W, Yang, K, Chen, C, Wu, C, Chi, H, Mu, Z, Wang, Z, Yang, H. | | Deposit date: | 2016-04-29 | | Release date: | 2016-05-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.796 Å) | | Cite: | The crystal structure of Zika virus helicase: basis for antiviral drug design
Protein Cell, 7, 2016
|
|
1EGA
 
 | | CRYSTAL STRUCTURE OF A WIDELY CONSERVED GTPASE ERA | | Descriptor: | PROTEIN (GTP-BINDING PROTEIN ERA), SULFATE ION | | Authors: | Chen, X, Ji, X. | | Deposit date: | 1998-12-01 | | Release date: | 1999-07-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of ERA: a GTPase-dependent cell cycle regulator containing an RNA binding motif.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
6AP9
 
 | |
4F7V
 
 | | Crystal structure of E. coli HPPK in complex with bisubstrate analogue inhibitor J1D (HP26) | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, 5'-{[2-({N-[(2-amino-7,7-dimethyl-4-oxo-3,4,7,8-tetrahydropteridin-6-yl)carbonyl]glycyl}amino)ethyl]sulfonyl}-5'-deoxyadenosine | | Authors: | Shaw, G, Shi, G, Ji, X. | | Deposit date: | 2012-05-16 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Bisubstrate analog inhibitors of 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase: New lead exhibits a distinct binding mode.
Bioorg.Med.Chem., 20, 2012
|
|
7WWK
 
 | | Local refinement of the SARS-CoV-2 BA.1 Spike trimer in complex with 55A8 Fab | | Descriptor: | 55A8 light chain, Spike glycoprotein | | Authors: | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | Deposit date: | 2022-02-13 | | Release date: | 2023-02-15 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Local refinement of the SARS-CoV-2 BA.1 Spike trimer in complex with 55A8 Fab
To Be Published
|
|
7WWJ
 
 | | SARS-CoV-2 BA.1 Spike trimer in complex with 55A8 Fab in the class 2 conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 55A8 heavy chain, ... | | Authors: | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | Deposit date: | 2022-02-13 | | Release date: | 2023-02-15 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | EM structure of SARS-CoV-2 Omicron variant spike glycoprotein and 55A8
To Be Published
|
|
7WWI
 
 | | SARS-CoV-2 BA.1 Spike trimer in complex with 55A8 Fab in the class 1 conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 55A8 heavy chain, ... | | Authors: | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | Deposit date: | 2022-02-13 | | Release date: | 2023-02-15 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | SARS-CoV-2 BA.1 Spike trimer in complex with 55A8 Fab in the class 1 conformation
To Be Published
|
|
7XJ9
 
 | | Local refinement of the SARS-CoV-2 BA.1 Spike trimer in complex with 55A8 Fab and 58G6 Fab | | Descriptor: | 55A8 heavy chain, 55A8 light chain, 58G6 heavy chain, ... | | Authors: | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | Deposit date: | 2022-04-15 | | Release date: | 2023-04-19 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Local refinement of the SARS-CoV-2 BA.1 Spike trimer in complex with 55A8 Fab and 58G6 Fab
To Be Published
|
|
7XJ8
 
 | | SARS-CoV-2 BA.1 Spike trimer in complex with 55A8 Fab and 58G6 Fab in the class 2 conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 55A8 heavy chain, ... | | Authors: | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | Deposit date: | 2022-04-15 | | Release date: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | SARS-CoV-2 BA.1 Spike trimer in complex with 55A8 Fab and 58G6 Fab in the class 2 conformation
To Be Published
|
|
7XJ6
 
 | | SARS-CoV-2 BA.1 Spike trimer in complex with 55A8 Fab and 58G6 Fab in the class 1 conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 55A8 heavy chain, ... | | Authors: | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | Deposit date: | 2022-04-15 | | Release date: | 2023-04-19 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | SARS-CoV-2 BA.1 Spike trimer in complex with 55A8 Fab and 58G6 Fab in the class 1 conformation
To Be Published
|
|
2G75
 
 | | Crystal Structure of anti-SARS m396 Antibody | | Descriptor: | IGG Heavy Chain, IGG Light Chain | | Authors: | Prabakaran, P, Gan, J.H, Feng, Y, Zhu, Z.Y, Xiao, X.D, Ji, X, Dimitrov, D.S. | | Deposit date: | 2006-02-27 | | Release date: | 2006-04-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structure of severe acute respiratory syndrome coronavirus receptor-binding domain complexed with neutralizing antibody.
J.Biol.Chem., 281, 2006
|
|
1XCA
 
 | | APO-CELLULAR RETINOIC ACID BINDING PROTEIN II | | Descriptor: | CELLULAR RETINOIC ACID BINDING PROTEIN TYPE II | | Authors: | Chen, X, Ji, X. | | Deposit date: | 1996-12-31 | | Release date: | 1998-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of apo-cellular retinoic acid-binding protein type II (R111M) suggests a mechanism of ligand entry.
J.Mol.Biol., 278, 1998
|
|
6AN4
 
 | | Crystal structure of Escherichia coli HPPK in complex with bisubstrate analogue inhibitor HP-39 (J1F) | | Descriptor: | ((2-(2-amino-7,7-dimethyl-4-oxo-3,4,7,8-tetrahydropteridine-6-carboxamido)-N-(2-((((2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl)methyl)amino)-2-oxoethyl)acetamido)methyl)phosphonic acid, 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, CHLORIDE ION | | Authors: | Shaw, G.X, Shi, G, Ji, X. | | Deposit date: | 2017-08-12 | | Release date: | 2018-08-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Bisubstrate analog inhibitors of HPPK: Transition state mimetics
to be published
|
|
2QX0
 
 | | Crystal Structure of Yersinia pestis HPPK (Ternary Complex) | | Descriptor: | 2-AMINO-6-HYDROXYMETHYL-7,8-DIHYDRO-3H-PTERIDIN-4-ONE, 7,8-dihydro-6-hydroxymethylpterin-pyrophosphokinase, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, ... | | Authors: | Blaszczyk, J, Cherry, S, Tropea, J.E, Waugh, D.S, Ji, X. | | Deposit date: | 2007-08-10 | | Release date: | 2007-10-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and activity of Yersinia pestis 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase as a novel target for the development of antiplague therapeutics.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
3UDV
 
 | | Crystal structure of E. coli HPPK in complex with bisubstrate analogue inhibitor J1C | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, 5'-S-[1-(2-{[(2-amino-7,7-dimethyl-4-oxo-3,4,7,8-tetrahydropteridin-6-yl)carbonyl]amino}ethyl)piperidin-4-yl]-5'-thioadenosine, ACETATE ION | | Authors: | Shaw, G, Shi, G, Ji, X. | | Deposit date: | 2011-10-28 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Bisubstrate analogue inhibitors of 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase: New design with improved properties.
Bioorg.Med.Chem., 20, 2012
|
|
