4CBH
 
 | | Pestivirus NS3 helicase | | Descriptor: | SERINE PROTEASE NS3 | | Authors: | Tortorici, M.A, Duquerroy, S, Kwok, J, Vonrhein, C, Perez, J, Lamp, B, Bricogne, G, Rumenapf, T, Vachette, P, Rey, F.A. | | Deposit date: | 2013-10-14 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | X-Ray Structure of the Pestivirus Ns3 Helicase and its Conformation in Solution.
J.Virol., 89, 2015
|
|
4CBM
 
 | | Pestivirus NS3 helicase | | Descriptor: | SERINE PROTEASE NS3 | | Authors: | Tortorici, M.A, Duquerroy, S, Kwok, J, Vonrhein, C, Perez, J, Lamp, B, Bricogne, G, Rumenapf, T, Vachette, P, Rey, F.A. | | Deposit date: | 2013-10-14 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | X-Ray Structure of the Pestivirus Ns3 Helicase and its Conformation in Solution.
J.Virol., 89, 2015
|
|
6Z48
 
 | | Crystal structure of Thrombin in complex with macrocycle X1vE | | Descriptor: | 5-chloranyl-N-[[(4S,15R)-2,5,13,16-tetrakis(oxidanylidene)-15-propan-2-yl-9,10-dithia-3,6,14,17-tetrazabicyclo[17.3.1]tricosa-1(22),19(23),20-trien-4-yl]methyl]thiophene-2-carboxamide, SODIUM ION, Thrombin heavy chain, ... | | Authors: | Angelini, A, Habeshian, S, Heinis, C, Cendron, L. | | Deposit date: | 2020-05-23 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Synthesis and direct assay of large macrocycle diversities by combinatorial late-stage modification at picomole scale.
Nat Commun, 13, 2022
|
|
6GWE
 
 | | Crystal structure of Thrombin bound to P2 macrocycle | | Descriptor: | (10S,14S,17R)-14-(3-carbamimidamidopropyl)-3-[[2-(hydroxymethyl)phenyl]methyl]-5,12,15-tris(oxidanylidene)-19-thia-3,6,13,16-tetrazatricyclo[19.4.0.0^{6,10}]pentacosa-1(21),22,24-triene-17-carboxamide, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cendron, L, Angelini, A, Kale, S.S, Bergeron-Brlek, M, Wu, Y, Heinis, C. | | Deposit date: | 2018-06-23 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Thiol-to-amine cyclization reaction enables screening of large libraries of macrocyclic compounds and the generation of sub-kilodalton ligands.
Sci Adv, 5, 2019
|
|
8ASE
 
 | | Crystal structure of Thrombin in complex with macrocycle T3 | | Descriptor: | (8~{S},14~{S},18~{E})-8-[(4-chlorophenyl)methyl]-3,21-dithia-7,10,16-triazatricyclo[21.2.2.1^{10,14}]octacosa-1(26),18,23(27),24-tetraene-6,9,15-trione, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chinellato, M, Angelini, A, Nielsen, A, Heinis, C, Cendron, L. | | Deposit date: | 2022-08-19 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of Thrombin in complex with optimized macrocycles T1 and T3
To Be Published
|
|
8ASF
 
 | | Crystal structure of Thrombin in complex with macrocycle T1 | | Descriptor: | 5-chloranyl-~{N}-[[(9~{S},15~{R})-8,14,17-tris(oxidanylidene)-3,20-dithia-7,13,16-triazatetracyclo[20.2.2.1^{5,7}.1^{9,13}]octacosa-1(25),22(26),23-trien-15-yl]methyl]thiophene-2-carboxamide, Thrombin heavy chain, Thrombin light chain | | Authors: | Chinellato, M, Angelini, A, Nielsen, A, Heinis, C, Cendron, L. | | Deposit date: | 2022-08-19 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Crystal structure of Thrombin in complex with optimized macrocycles T1 and T3
To Be Published
|
|
2WIT
 
 | | CRYSTAL STRUCTURE OF THE SODIUM-COUPLED GLYCINE BETAINE SYMPORTER BETP FROM CORYNEBACTERIUM GLUTAMICUM WITH BOUND SUBSTRATE | | Descriptor: | GLYCINE BETAINE TRANSPORTER BETP, TRIMETHYL GLYCINE | | Authors: | Ressl, S, Terwisscha Van Scheltinga, A.C, Vonrhein, C, Ott, V, Ziegler, C. | | Deposit date: | 2009-05-17 | | Release date: | 2009-05-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Molecular Basis of Transport and Regulation in the Na(+)/Betaine Symporter Betp.
Nature, 458, 2009
|
|
2V4J
 
 | | THE CRYSTAL STRUCTURE OF Desulfovibrio vulgaris DISSIMILATORY SULFITE REDUCTASE BOUND TO DsrC PROVIDES NOVEL INSIGHTS INTO THE MECHANISM OF SULFATE RESPIRATION | | Descriptor: | 3,3',3'',3'''-[(1R,2S,3S,4S,7S,8S,11S,12S,13S,16S,19S)-3,8,13,17-tetrakis(carboxylatomethyl)-8,13-dimethyl-1,2,3,4,7,8,11,12,13,16,19,20,22,24-tetradecahydroporphyrin-2,7,12,18-tetrayl]tetrapropanoate, IRON/SULFUR CLUSTER, SIROHEME, ... | | Authors: | Oliveira, T.F, Vonrhein, C, Matias, P.M, Venceslau, S.S, Pereira, I.A.C, Archer, M. | | Deposit date: | 2008-09-22 | | Release date: | 2008-12-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of Desulfovibrio Vulgaris Dissimilatory Sulfite Reductase Bound to Dsrc Provides Novel Insights Into the Mechanism of Sulfate Respiration.
J.Biol.Chem., 283, 2008
|
|
2WJ8
 
 | | Respiratory Syncitial Virus RiboNucleoProtein | | Descriptor: | BORATE ION, NUCLEOPROTEIN, RNA (5'-R(*CP*CP*CP*CP*CP*C)-3') | | Authors: | Tawar, R.G, Duquerroy, S, Vonrhein, C, Varela, P.F, Damier-Piolle, L, Castagne, N, MacLellan, K, Bedouelle, H, Bricogne, G, Bhella, D, Eleouet, J, Rey, F.A. | | Deposit date: | 2009-05-25 | | Release date: | 2009-12-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Crystal Structure of a Nucleocapsid-Like Nucleoprotein-RNA Complex of Respiratory Syncytial Virus
Science, 326, 2009
|
|
5NFY
 
 | | SARS-CoV nsp10/nsp14 dynamic complex | | Descriptor: | DI(HYDROXYETHYL)ETHER, Polyprotein 1ab, ZINC ION | | Authors: | Ferron, F, Gluais, L, Vonrhein, C, Bricogne, G, Canard, B, Imbert, I. | | Deposit date: | 2017-03-16 | | Release date: | 2018-01-10 | | Last modified: | 2019-02-20 | | Method: | X-RAY DIFFRACTION (3.382 Å) | | Cite: | Structural and molecular basis of mismatch correction and ribavirin excision from coronavirus RNA.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2W2G
 
 | | Human SARS coronavirus unique domain | | Descriptor: | NON-STRUCTURAL PROTEIN 3, SULFATE ION | | Authors: | Tan, J, Vonrhein, C, Smart, O.S, Bricogne, G, Bollati, M, Kusov, Y, Hansen, G, Mesters, J.R, Schmidt, C.L, Hilgenfeld, R. | | Deposit date: | 2008-10-30 | | Release date: | 2009-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | The Sars-Unique Domain (Sud) of Sars Coronavirus Contains Two Macrodomains that Bind G-Quadruplexes.
Plos Pathog., 5, 2009
|
|
2VKZ
 
 | | Structure of the cerulenin-inhibited fungal fatty acid synthase type I multienzyme complex | | Descriptor: | (2S, 3R)-3-HYDROXY-4-OXO-7,10-TRANS,TRANS-DODECADIENAMIDE, FATTY ACID SYNTHASE SUBUNIT ALPHA, ... | | Authors: | Johansson, P, Wiltschi, B, Kumari, P, Kessler, B, Vonrhein, C, Vonck, J, Oesterhelt, D, Grininger, M. | | Deposit date: | 2008-01-07 | | Release date: | 2008-08-12 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Inhibition of the Fungal Fatty Acid Synthase Type I Multienzyme Complex.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2WCT
 
 | | human SARS coronavirus unique domain (triclinic form) | | Descriptor: | NON-STRUCTURAL PROTEIN 3 | | Authors: | Tan, J, Vonrhein, C, Smart, O.S, Bricogne, G, Bollati, M, Hansen, G, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2009-03-16 | | Release date: | 2009-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | The Sars-Unique Domain (Sud) of Sars Coronavirus Contains Two Macrodomains that Bind G-Quadruplexes.
Plos Pathog., 5, 2009
|
|
5J9P
 
 | | KcsA in vitro | | Descriptor: | Fab, POTASSIUM ION, pH-gated potassium channel KcsA | | Authors: | Matulef, K, Valiyaveetil, F.I. | | Deposit date: | 2016-04-10 | | Release date: | 2016-07-20 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Combining in Vitro Folding with Cell Free Protein Synthesis for Membrane Protein Expression.
Biochemistry, 55, 2016
|
|
3SJD
 
 | | Crystal structure of S. cerevisiae Get3 with bound ADP-Mg2+ in complex with Get2 cytosolic domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATPase GET3, Golgi to ER traffic protein 2, ... | | Authors: | Reitz, S, Wild, K, Sinning, I. | | Deposit date: | 2011-06-21 | | Release date: | 2011-07-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.6 Å) | | Cite: | Structural basis for tail-anchored membrane protein biogenesis by the Get3-receptor complex.
Science, 333, 2011
|
|
3SJB
 
 | | Crystal structure of S. cerevisiae Get3 in the open state in complex with Get1 cytosolic domain | | Descriptor: | ATPase GET3, Golgi to ER traffic protein 1, PHOSPHATE ION, ... | | Authors: | Reitz, S, Wild, K, Sinning, I. | | Deposit date: | 2011-06-21 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for tail-anchored membrane protein biogenesis by the Get3-receptor complex.
Science, 333, 2011
|
|
3SJA
 
 | | Crystal structure of S. cerevisiae Get3 in the open state in complex with Get1 cytosolic domain | | Descriptor: | ATPase GET3, Golgi to ER traffic protein 1, PHOSPHATE ION, ... | | Authors: | Reitz, S, Wild, K, Sinning, I. | | Deposit date: | 2011-06-21 | | Release date: | 2011-07-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for tail-anchored membrane protein biogenesis by the Get3-receptor complex.
Science, 333, 2011
|
|
3SJC
 
 | |
4CSR
 
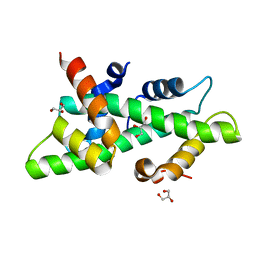 | | High resolution crystal structure of the histone fold dimer (NF-YB)-(NF-YC) | | Descriptor: | GLYCEROL, NUCLEAR TRANSCRIPTION FACTOR Y SUBUNIT BETA, NUCLEAR TRANSCRIPTION FACTOR Y SUBUNIT GAMMA | | Authors: | Gnesutta, N, Cocolo, S, Mantovani, R, Bolognesi, M, Nardini, M. | | Deposit date: | 2014-03-09 | | Release date: | 2015-03-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High Resolution Crystal Structure of the Histone Fold Dimer (NF-Yb)-(NF-Yc)
To be Published
|
|
2XFB
 
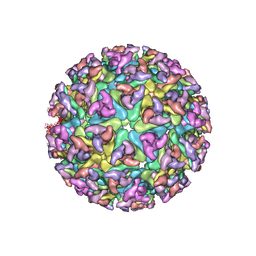 | | CHIKUNGUNYA E1 E2 ENVELOPE GLYCOPROTEINS FITTED IN SINDBIS VIRUS cryo- EM MAP | | Descriptor: | E1 ENVELOPE GLYCOPROTEIN, E2 ENVELOPE GLYCOPROTEIN | | Authors: | Voss, J.E, Vaney, M.C, Duquerroy, S, Rey, F.A. | | Deposit date: | 2010-05-21 | | Release date: | 2010-11-24 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Glycoprotein Organization of Chikungunya Virus Particles Revealed by X-Ray Crystallography
Nature, 468, 2010
|
|
2XFC
 
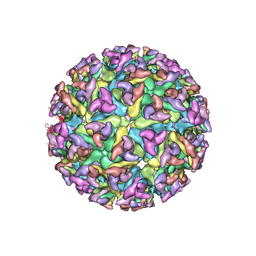 | | CHIKUNGUNYA E1 E2 ENVELOPE GLYCOPROTEINS FITTED IN SEMLIKI FOREST VIRUS cryo-EM MAP | | Descriptor: | E1 ENVELOPE GLYCOPROTEIN, E2 ENVELOPE GLYCOPROTEIN | | Authors: | Voss, J.E, Vaney, M.C, Duquerroy, S, Rey, F.A. | | Deposit date: | 2010-05-21 | | Release date: | 2010-11-24 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Glycoprotein Organization of Chikungunya Virus Particles Revealed by X-Ray Crystallography
Nature, 468, 2010
|
|
1HNW
 
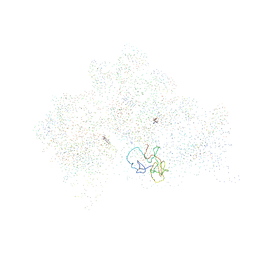 | | STRUCTURE OF THE THERMUS THERMOPHILUS 30S RIBOSOMAL SUBUNIT IN COMPLEX WITH TETRACYCLINE | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Brodersen, D.E, Clemons Jr, W.M, Carter, A.P, Morgan-Warren, R, Wimberly, B.T, Ramakrishnan, V. | | Deposit date: | 2000-12-08 | | Release date: | 2001-02-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit.
Cell(Cambridge,Mass.), 103, 2000
|
|
1HR0
 
 | | CRYSTAL STRUCTURE OF INITIATION FACTOR IF1 BOUND TO THE 30S RIBOSOMAL SUBUNIT | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Carter, A.P, Clemons Jr, W.M, Brodersen, D.E, Morgan-Warren, R.J, Wimberly, B.T, Ramakrishnan, V. | | Deposit date: | 2000-12-20 | | Release date: | 2001-01-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of an initiation factor bound to the 30S ribosomal subunit.
Science, 291, 2001
|
|
8Q4S
 
 | |
1X18
 
 | | Contact sites of ERA GTPase on the THERMUS THERMOPHILUS 30S SUBUNIT | | Descriptor: | 30S ribosomal protein S11, 30S ribosomal protein S18, 30S ribosomal protein S2, ... | | Authors: | Sharma, M.R, Barat, C, Agrawal, R.K. | | Deposit date: | 2005-04-02 | | Release date: | 2005-05-17 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (13.5 Å) | | Cite: | Interaction of Era with the 30S Ribosomal Subunit Implications for 30S Subunit Assembly
Mol.Cell, 18, 2005
|
|
