1BEU
 
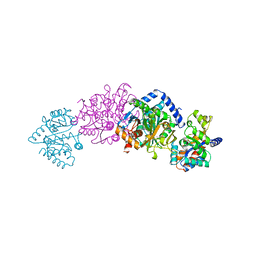 | | TRP SYNTHASE (D60N-IPP-SER) WITH K+ | | Descriptor: | INDOLE-3-PROPANOL PHOSPHATE, POTASSIUM ION, TRYPTOPHAN SYNTHASE, ... | | Authors: | Rhee, S, Mozzarelli, A, Miles, E.W, Davies, D.R. | | Deposit date: | 1998-05-18 | | Release date: | 1998-08-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cryocrystallography and microspectrophotometry of a mutant (alpha D60N) tryptophan synthase alpha 2 beta 2 complex reveals allosteric roles of alpha Asp60.
Biochemistry, 37, 1998
|
|
6Q0D
 
 | | CRYSTAL STRUCTURE OF LDHA IN COMPLEX WITH COMPOUND NCGC00384414-01 AT 2.05 A RESOLUTION | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2-{3-[3-(cyclopentylethynyl)-4-fluorophenyl]-5-(cyclopropylmethyl)-4-[(3-fluoro-4-sulfamoylphenyl)methyl]-1H-pyrazol-1-yl}-1,3-thiazole-4-carboxylic acid, GLYCEROL, ... | | Authors: | Dranow, D.M, Davies, D.R. | | Deposit date: | 2019-08-01 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Pyrazole-Based Lactate Dehydrogenase Inhibitors with Optimized Cell Activity and Pharmacokinetic Properties.
J.Med.Chem., 63, 2020
|
|
4APR
 
 | |
1ITG
 
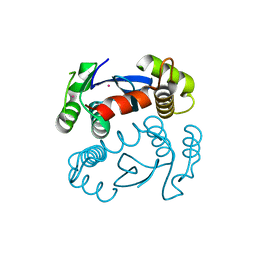 | | CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF HIV-1 INTEGRASE: SIMILARITY TO OTHER POLYNUCLEOTIDYL TRANSFERASES | | Descriptor: | CACODYLATE ION, HIV-1 INTEGRASE | | Authors: | Dyda, F, Hickman, A.B, Jenkins, T.M, Engelman, A, Craigie, R, Davies, D.R. | | Deposit date: | 1994-11-21 | | Release date: | 1995-05-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases.
Science, 266, 1994
|
|
1HQO
 
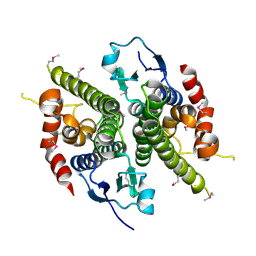 | | CRYSTAL STRUCTURE OF THE NITROGEN REGULATION FRAGMENT OF THE YEAST PRION PROTEIN URE2P | | Descriptor: | URE2 PROTEIN | | Authors: | Umland, T.C, Taylor, K.L, Rhee, S, Wickner, R.B, Davies, D.R. | | Deposit date: | 2000-12-18 | | Release date: | 2001-02-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of the nitrogen regulation fragment of the yeast prion protein Ure2p.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
2TRS
 
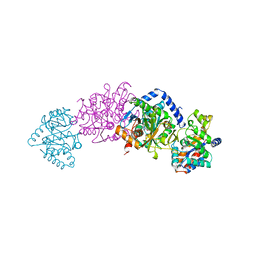 | | CRYSTAL STRUCTURES OF MUTANT (BETAK87T) TRYPTOPHAN SYNTHASE ALPHA2 BETA2 COMPLEX WITH LIGANDS BOUND TO THE ACTIVE SITES OF THE ALPHA AND BETA SUBUNITS REVEAL LIGAND-INDUCED CONFORMATIONAL CHANGES | | Descriptor: | INDOLE-3-PROPANOL PHOSPHATE, SODIUM ION, TRYPTOPHAN SYNTHASE, ... | | Authors: | Rhee, S, Parris, K.D, Hyde, C.C, Ahmed, S.A, Miles, E.W, Davies, D.R. | | Deposit date: | 1997-01-07 | | Release date: | 1997-04-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal structures of a mutant (betaK87T) tryptophan synthase alpha2beta2 complex with ligands bound to the active sites of the alpha- and beta-subunits reveal ligand-induced conformational changes.
Biochemistry, 36, 1997
|
|
2TSY
 
 | | CRYSTAL STRUCTURES OF MUTANT (BETAK87T) TRYPTOPHAN SYNTHASE ALPHA2 BETA2 COMPLEX WITH LIGANDS BOUND TO THE ACTIVE SITES OF THE ALPHA AND BETA SUBUNITS REVEAL LIGAND-INDUCED CONFORMATIONAL CHANGES | | Descriptor: | SN-GLYCEROL-3-PHOSPHATE, SODIUM ION, TRYPTOPHAN SYNTHASE, ... | | Authors: | Rhee, S, Parris, K.D, Hyde, C.C, Ahmed, S.A, Miles, E.W, Davies, D.R. | | Deposit date: | 1997-01-03 | | Release date: | 1997-04-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of a mutant (betaK87T) tryptophan synthase alpha2beta2 complex with ligands bound to the active sites of the alpha- and beta-subunits reveal ligand-induced conformational changes.
Biochemistry, 36, 1997
|
|
2TYS
 
 | | CRYSTAL STRUCTURES OF MUTANT (BETAK87T) TRYPTOPHAN SYNTHASE ALPHA2 BETA2 COMPLEX WITH LIGANDS BOUND TO THE ACTIVE SITES OF THE ALPHA AND BETA SUBUNITS REVEAL LIGAND-INDUCED CONFORMATIONAL CHANGES | | Descriptor: | SODIUM ION, TRYPTOPHAN SYNTHASE, [3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-L-TRYPTOPHANE | | Authors: | Rhee, S, Parris, K.D, Hyde, C.C, Ahmed, S.A, Miles, E.W, Davies, D.R. | | Deposit date: | 1997-01-08 | | Release date: | 1997-04-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of a mutant (betaK87T) tryptophan synthase alpha2beta2 complex with ligands bound to the active sites of the alpha- and beta-subunits reveal ligand-induced conformational changes.
Biochemistry, 36, 1997
|
|
1A5A
 
 | | CRYO-CRYSTALLOGRAPHY OF A TRUE SUBSTRATE, INDOLE-3-GLYCEROL PHOSPHATE, BOUND TO A MUTANT (ALPHAD60N) TRYPTOPHAN SYNTHASE ALPHA2BETA2 COMPLEX REVEALS THE CORRECT ORIENTATION OF ACTIVE SITE ALPHA GLU 49 | | Descriptor: | POTASSIUM ION, PYRIDOXAL-5'-PHOSPHATE, TRYPTOPHAN SYNTHASE (ALPHA CHAIN), ... | | Authors: | Rhee, S, Miles, E.W, Davies, D.R. | | Deposit date: | 1998-02-12 | | Release date: | 1998-05-27 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cryo-crystallography of a true substrate, indole-3-glycerol phosphate, bound to a mutant (alphaD60N) tryptophan synthase alpha2beta2 complex reveals the correct orientation of active site alphaGlu49.
J.Biol.Chem., 273, 1998
|
|
1A30
 
 | | HIV-1 PROTEASE COMPLEXED WITH A TRIPEPTIDE INHIBITOR | | Descriptor: | HIV-1 PROTEASE, TRIPEPTIDE GLU-ASP-LEU | | Authors: | Louis, J.M, Dyda, F, Nashed, N.T, Kimmel, A.R, Davies, D.R. | | Deposit date: | 1998-01-27 | | Release date: | 1998-04-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Hydrophilic peptides derived from the transframe region of Gag-Pol inhibit the HIV-1 protease.
Biochemistry, 37, 1998
|
|
1A5B
 
 | | CRYO-CRYSTALLOGRAPHY OF A TRUE SUBSTRATE, INDOLE-3-GLYCEROL PHOSPHATE, BOUND TO A MUTANT (ALPHA D60N) TRYPTOPHAN SYNTHASE ALPHA2BETA2 COMPLEX REVEALS THE CORRECT ORIENTATION OF ACTIVE SITE ALPHA GLU 49 | | Descriptor: | INDOLE-3-GLYCEROL PHOSPHATE, POTASSIUM ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Rhee, S, Miles, E.W, Davies, D.R. | | Deposit date: | 1998-02-12 | | Release date: | 1998-05-27 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cryo-crystallography of a true substrate, indole-3-glycerol phosphate, bound to a mutant (alphaD60N) tryptophan synthase alpha2beta2 complex reveals the correct orientation of active site alphaGlu49.
J.Biol.Chem., 273, 1998
|
|
1QS4
 
 | | Core domain of HIV-1 integrase complexed with Mg++ and 1-(5-chloroindol-3-yl)-3-hydroxy-3-(2H-tetrazol-5-yl)-propenone | | Descriptor: | 1-(5-CHLOROINDOL-3-YL)-3-HYDROXY-3-(2H-TETRAZOL-5-YL)-PROPENONE, MAGNESIUM ION, PROTEIN (HIV-1 INTEGRASE (E.C.2.7.7.49)) | | Authors: | Goldgur, Y, Craigie, R, Fujiwara, T, Yoshinaga, T, Davies, D.R. | | Deposit date: | 1999-06-25 | | Release date: | 1999-11-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the HIV-1 integrase catalytic domain complexed with an inhibitor: a platform for antiviral drug design.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1YRI
 
 | | Chicken villin subdomain HP-35, N68H, pH6.4 | | Descriptor: | ACETATE ION, IODIDE ION, Villin | | Authors: | Chiu, T.K, Kubelka, J, Herbst-Irmer, R, Eaton, W.A, Hofrichter, J, Davies, D.R. | | Deposit date: | 2005-02-03 | | Release date: | 2005-05-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | High-resolution x-ray crystal structures of the villin headpiece subdomain, an ultrafast folding protein.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1YRF
 
 | | Chicken villin subdomain HP-35, N68H, pH6.7 | | Descriptor: | ACETATE ION, SULFATE ION, Villin | | Authors: | Chiu, T.K, Kubelka, J, Herbst-Irmer, R, Eaton, W.A, Hofrichter, J, Davies, D.R. | | Deposit date: | 2005-02-03 | | Release date: | 2005-05-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | High-resolution x-ray crystal structures of the villin headpiece subdomain, an ultrafast folding protein.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1YQV
 
 | | The crystal structure of the antibody Fab HyHEL5 complex with lysozyme at 1.7A resolution | | Descriptor: | Hen Egg White Lysozyme, HyHEL-5 Antibody Heavy Chain, HyHEL-5 Antibody Light Chain | | Authors: | Cohen, G.H, Silverton, E.W, Padlan, E.A, Dyda, F, Wibbenmeyer, J.A, Wilson, R.C, Davies, D.R. | | Deposit date: | 2005-02-02 | | Release date: | 2005-04-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Water molecules in the antibody-antigen interface of the structure of the Fab HyHEL-5-lysozyme complex at 1.7 A resolution: comparison with results from isothermal titration calorimetry.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1ZYM
 
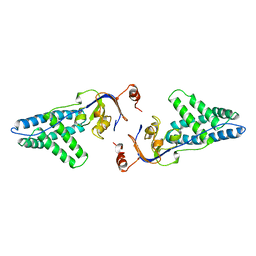 | | AMINO TERMINAL DOMAIN OF ENZYME I FROM ESCHERICHIA COLI | | Descriptor: | ENZYME I | | Authors: | Liao, D.-I, Davies, D.R. | | Deposit date: | 1996-05-21 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The first step in sugar transport: crystal structure of the amino terminal domain of enzyme I of the E. coli PEP: sugar phosphotransferase system and a model of the phosphotransfer complex with HPr.
Structure, 4, 1996
|
|
2A0Z
 
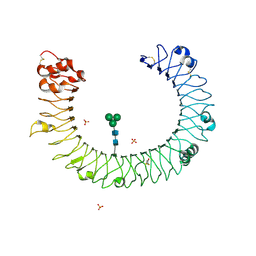 | | The molecular structure of toll-like receptor 3 ligand binding domain | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bell, J.K, Botos, I, Hall, P.R, Askins, J, Shiloach, J, Segal, D.M, Davies, D.R. | | Deposit date: | 2005-06-17 | | Release date: | 2005-08-02 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The molecular structure of the Toll-like receptor 3 ligand-binding domain
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
3CIG
 
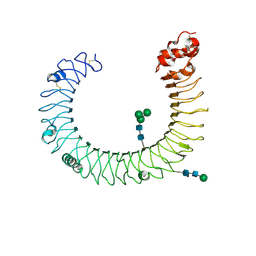 | | Crystal structure of mouse TLR3 ectodomain | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Liu, L, Botos, I, Wang, Y, Leonard, J.N, Shiloach, J, Segal, D.M, Davies, D.R. | | Deposit date: | 2008-03-11 | | Release date: | 2008-05-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Structural basis of toll-like receptor 3 signaling with double-stranded RNA.
Science, 320, 2008
|
|
4NPC
 
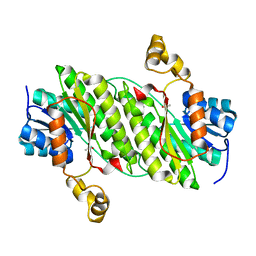 | | Crystal Structure of an Oxidoreductase, Short-Chain Dehydrogenase/Reductase Family Protein from Brucella suis | | Descriptor: | ACETATE ION, Sorbitol dehydrogenase | | Authors: | Dranow, D.M, Davies, D.R, Edwards, T.E, Lorimer, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2013-11-21 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of an Oxidoreductase, Short-Chain Dehydrogenase/Reductase Family Protein from Brucella suis
To be Published
|
|
1WY4
 
 | | Chicken villin subdomain HP-35, K65(NLE), N68H, pH5.1 | | Descriptor: | IODIDE ION, SODIUM ION, Villin | | Authors: | Chiu, T.K, Kubelka, J, Herbst-Irmer, R, Eaton, W.A, Hofrichter, J, Davies, D.R. | | Deposit date: | 2005-02-04 | | Release date: | 2005-05-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | High-resolution x-ray crystal structures of the villin headpiece subdomain, an ultrafast folding protein.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1WY3
 
 | | Chicken villin subdomain HP-35, K65(NLE), N68H, pH7.0 | | Descriptor: | Villin | | Authors: | Chiu, T.K, Kubelka, J, Herbst-Irmer, R, Eaton, W.A, Hofrichter, J, Davies, D.R. | | Deposit date: | 2005-02-04 | | Release date: | 2005-05-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | High-resolution x-ray crystal structures of the villin headpiece subdomain, an ultrafast folding protein.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1D3R
 
 | | CRYSTAL STRUCTURE OF TRIPLEX DNA | | Descriptor: | DNA (5'-D(*CP*(BRU)P*CP*CP*(BRU)P*CP*CP*GP*CP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*GP*CP*GP*GP*AP*G)-3') | | Authors: | Rhee, S, Han, Z.-J, Liu, K, Todd Miles, H.T, Davies, D.R. | | Deposit date: | 1999-09-30 | | Release date: | 2000-01-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a triple helical DNA with a triplex-duplex junction.
Biochemistry, 38, 1999
|
|
1BIU
 
 | | HIV-1 INTEGRASE CORE DOMAIN COMPLEXED WITH MG++ | | Descriptor: | HIV-1 INTEGRASE, MAGNESIUM ION | | Authors: | Goldgur, Y, Dyda, F, Hickman, A.B, Jenkins, T.M, Craigie, R, Davies, D.R. | | Deposit date: | 1998-06-19 | | Release date: | 1998-08-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three new structures of the core domain of HIV-1 integrase: an active site that binds magnesium.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1CI4
 
 | |
1DHS
 
 | |
